
The long intergenic non-protein coding RNA 472 (LINC00472) aggravates neuropathic pain through the microRNA-300/high mobility group box protein 1 axis: a study using the chronic constrictive injury rat model
Introduction
Neuropathic pain is caused by the injury or dysfunction of the somatic nervous system. It has a high incidence in the 40–60-year-old age group and in the elderly (1,2), and is characterized by hyperalgesia, abnormal headaches, and hypoalgesia (3). The pathogenesis of neuropathic pain is complex, and treatment is difficult. Recently, the neuroimmune mechanism has been widely recognized by researchers. Immune cells and glial cells are activated in the state of neuropathic pain and communicate with neurons by releasing a large number of inflammatory mediators (such as cytokines, chemokines, etc.). These inflammatory mediators can regulate inflammatory response and pain-related signal transduction by interacting with neurotransmitters and their receptors. Thus it plays a vital role in the occurrence and development of neuropathic pain (4,5). Currently, the first line treatment for neuropathic pain includes analgesics, antidepressants, and NMDA antagonists. However, the clinical efficacy is unclear and thus, their clinical application is limited (6). Therefore, it is crucial to understand the mechanisms underlying neuropathic pain so as to develop effective prevention and treatment strategies.
Long non-coding RNAs (lncRNAs) are important RNA molecules located in the nucleus or cytoplasm of the cell. The expression of lncRNAs is more specific to the protein coding gene than to a specific tissue or cell type (7). LncRNAs can participate in post transcriptional gene regulation as well as RNA maturation and transport by controlling protein synthesis and transcriptional gene silencing by modifying chromatin structure (8). Studies on gene knockout animal models have shown that a variety of lncRNAs may play important roles in nervous system functions and the onset of numerous diseases. Indeed, there has been some experimental evidence to support the development therapies that target lncRNAs (9,10). Liu et al. showed that lnc-DILC (lncRNA downregulated in liver cancer stem cells) exacerbated neuropathic pain by activating the JAK2/STAT3 pathway via suppressor of cytokine signaling 3 (SOC3) (11). Similarly, upregulating the lncRNA LOC100911498 promoted neuropathic pain and nerve injury (12). The long intergenic non-protein coding RNA 472 (LINC00472) has been shown to play an important role in the progression of cancer (13,14). In addition, studies have shown that LINC00472 aggravates lipopolysaccharide (LPS)-induced inflammatory responses by regulating the microRNA (miR)-373-3p/tripartite motif containing 8 (TRIM8) axis in acute liver injury (15). This suggested that LINC00472 is involved in the progression of inflammation. However, the regulatory role of LINC00472 on inflammation in neuropathic pain has not been fully elucidated.
MicroRNAs (miRNAs) are non-coding functional RNAs that negatively regulate polygene expression. Functional (or mature) miRNAs recognize and bind to certain homologous sequences in the 3'-untranslated regions (3'-UTRs) of mRNAs. To date, there have been many studies examining the application of miRNAs in the diagnosis and treatment of diseases (16,17). The regulation of miRNA-related gene expression is involved in the occurrence and development of learning and memory, neural remodeling, and neuronal degradation diseases (18). Studies have shown that miRNAs may play an important role in changing the expression of pain-related proteins. For example, studies have shown that miR-128-3p could alleviate neuropathic pain (19). Furthermore, miR-300 targets high mobility group box protein 1 (HMGB1) relieves cancer-induced neuropathic pain (20). However, the role of miR-300/HMGB1 in neuropathic pain is unclear. Other studies have also shown that downregulation of LINC00472 by targeting miR-128-3p and downregulation of aquaporin 4 (AQP4) regulated the progression of spinal cord traumatic neuropathic pain (21). The lncRNA MALAT1 has also been shown to promote the progression of neuropathic pain through the miR-154-5p/AQP9 axis (22). In addition, Zhang et al. showed that the downregulation of LINC00472 promoted the incidence of osteosarcoma by reducing FoxO1 expression through miR-300 (23). However, the precise role of LINC00472 in regulating miR-300 in neuropathic pain is unclear.
This study explored the roles and mechanisms of LINC00472 in neuropathic pain. Chronic constrictive injury (CCI) was established in rats as a model of neuropathic pain and the degree of pain and inflammation was assessed. The study demonstrated that the expression of LINC00472 was upregulated in CCI rats and miR-300 was the direct target of LINC00472. Further studies confirmed that the interaction between LINC00472, miR-300, and high mobility group box protein 1 (HMGB1) was associated with neuropathic pain. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/apm-21-2651).
Methods
Ethical statement
Animal experiments were approved by The First Affiliated Hospital of Southern University of Science and Technology Ethics Committee, and was compliance with the First Affiliated Hospital of Southern University of Science and Technology institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Construction of the chronic constrictive injury (CCI) model
Specific pathogen-free Sprague Dawley (SD) rats were purchased from Beijing Animal Research Institute. Ten SD rats, half male and half female, were about 9 weeks old. The weight of female was 240 g and that of male was 290–380 g. Rats were housed in a quiet room with good ventilation and air filtration, maintained at 20–25 °C and 50% relative humidity, with a 12 h:12 h light and dark cycle. Animals were given free access to water and food and bedding was changed daily. The rats adapted to the environment for 3 days prior to experiments. Animals were randomly divided into a sham operation group (n=5) and a model group (n=5). The CCI model was constructed as described previously (19). Briefly, rats underwent bilateral sciatic nerve ligation under pentobarbital (50–70 mg/kg) anesthesia. A 3 cm incision was made into the skin between the gluteal muscle and biceps femoris. The subcutaneous tissue and muscle membrane were passively separated, the trunk of sciatic nerve was exposed and a 7 mm long sciatic nerve was dissociated with a glass minute needle. Three loose knots with an interval of 1 mm were tied with 4-0 chromium catgut to ligate the sciatic nerve trunk until a short twitch was observed. In the sham operation group, the sciatic nerve was exposed and isolated, but not ligated.
Behavioral testing of the animal model
The mechanical pain threshold and the cold pain threshold were measured between 9:00–11:00 am every day for 3 consecutive days before the operation and again at days 0, 3, 7, 10, and 14 after the operation. To determine the mechanical pain threshold, rats were placed on a steel wire grid frame covered with a breathable plexiglass box for 10 minutes to adapt to the environment, and the mechanical pain threshold of the soles of both feet was measured with von Frey filaments. Each foot was measured three times and the average value was determined. To determination the cold pain threshold, a 1 mL syringe was used and 0.1 mL acetone was dropped on the plantar of the rats. Retraction of the foot indicated a positive response. Each plantar was assessed 5 times with an interval of 2 min and the positive rate (out of 10) for each rat was determined.
Cell culture and transfection
Rat pheochromocytoma PC12 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Dulbecco Modified Eagle Medium (DMEM; Gibco, Thermo Fisher Scientific, Inc., Waltham, Ma, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, USA), 100 units/mL penicillin, and 100 mg/mL streptomycin in a humidified environment of at 37 °C containing 5% CO2. The short hairpin RNA (shRNA) of LINC00472 and LINC00472 (RIBOOCO, Guangzhou, China), as well as miR-300, a miR-300 inhibitor, and a miR negative control were synthesized. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The total RNA from cells was extracted using TRIzol reagent (Thermo Fisher Scientific). The first strand of cDNA was synthesized by reverse transcription followed by amplification of the lipocalin-2 mRNA fragment using abi7300 fluorescence quantitative PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference gene and the relative value of the target gene was determined using the following equation: RQ =2−∆∆Ct. The primer sequences used in the experiment were all synthesized by Guangzhou Ruibo Biology Co., Ltd. β-actin was the standardized internal parameter of LINC00472 and HMGB1 mRNA, and U6 was the standardized internal parameter of miR-300.The gene specific primers were as follows: LINC00472 (forward: 5'-CAACACAACACAGGAGGGGA-3'; reverse: 5'-CCAAATAACGGGGGCTACCA-3'); miR-300 (forward: 5'-CCGGTTCAAGAGGAGACCAG-3'; reverse: 5'-GGAGGGTAATTGAGGAAGGA-3'); HMGB1 (forward: 5'-ACATCCAAAATCTTGATCAGTTA-3'; reverse: 5'-AGGACAGACTTTCAAAATGTTT-3'); β-actin (forward: 5'-TGAGAGGGAAATCGTGCGTGAC-3'; reverse: 5'-AAGAAGGAAGGCTGGAAAAGAG-3'); U6 (forward: 5'-CTCGCTTCGGCAGCACAT-3'; reverse: 5'-AACGCTTCACGAATTTGCGT-3').
Enzyme-linked immunosorbent assay (ELISA)
Protease inhibitors were added to cell lysates. The supernatant was centrifuged for 25 minutes at 4 °C at 14,000 g. The ELISA Kit (Invitrogen, USA) was used to detect the levels of interleukin-(IL)1β, IL-6, IL-4, IL-10, and tumor necrosis factor (TNF)-α.
Double luciferase experiments
The LINC00472 sequence was amplified and incorporated downstream of the luciferase gene in the pmirGLO plasmid (Promega, Fitchburg, WI, USA) to obtain pMIR-LINC00472-WT. To obtain a mutant structure (pMIR-LINC00472-MUT), a fragment containing a mutant target was designed. The pMIR-LINC00472-WT and pMIR-LINC00472-MUT, as well as miR-300 mimics and miR-negative control (NC), were co-transfected into human embryonic kidney HEK-293T cells with Lipofectamine 3000 (Thermo Fisher Scientific, Inc.). The Luciferase Reporter Assay System (Promega) was used to detect the luciferase activity 48 hours after transfection.
Western blot analysis
Total protein was extracted from cells and the protein concentration was measured using the BCA method. Protein samples were stored at −80 °C until use. Following protein denaturation, 20 µg protein/sample was loaded and separated by electrophoresis on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Samples were then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, Ma, USA) at a constant current of 300 mA. Membranes were blocked at room temperature with Tris-buffered saline solution with Tween 20 (TBST) containing 5% skimmed milk for 1 hour, followed by probing with the primary antibody anti-HMGB1 (ab18256, Abcam, Shanghai, China) at 4 °C overnight. Membranes were then washed with TBST 4 times for 8 min each time and incubated with the corresponding secondary antibody (1:2,000 dilution) for 1.5 h at room temperature. Following another 4 washes in TBST, protein bands were visualized by X-ray development using the Pierce ECL Western Blot Substrate Kit (Thermo Fisher Scientific).
RNA immunoprecipitation (RIP)
RIP was performed using the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, Ma, USA) according to the manufacturer’s instructions. Briefly, 2×107 cells were lysed and the resultant lysate was incubated with magnetic beads bound to normal mouse IgG (negative control) or human anti-ago2 antibody (Micropores). The immunoprecipitated RNA was extracted, and the bound target was confirmed by qRT-PCR and visualized via gel electrophoresis.
RNA pull-down assays
LINC00472 or antisense RNA was labeled with a biotin RNA labeling mixture (Roche, Mannhei, Germany) and incubated with streptavidin beads (Thermo, CA, USA) overnight at 4 °C. The mixture was then centrifuged at 3,000 g at 4 °C for 1 minute followed by 3 washes with washing buffer. A total of 2×107 cells were lysed and the resultant lysate was incubated with the bead biotin complexes at room temperature for 1 h. After washing with the washing buffer, the RNA bound to the beads was captured and extracted with Trizol for subsequent qPCR analysis.
Statistical analysis
All experiments were repeated at least 3 times and the data was analyzed using SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA). The mean ± standard deviation (x ± s) was used to describe the normal distribution of the measured data, the variance analysis was used to compare the measured data between groups, the Least Significant Difference (LSD) method was used to compare two groups, and the t test was used to compare the measured data between two groups. A P value <0.05 was considered statistically significant.
Results
LINC00472 participates in the development of neuropathic pain in the chronic constrictive injury (CCI) rat model
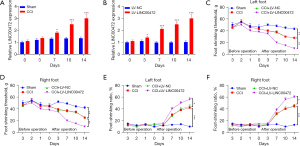
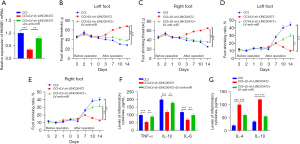
To determine the role of LINC00472 in neuropathic pain, a rat model of chronic constrictive injury (CCI) was constructed and the time-dependent expression of LINC00472 was assessed. In the CCI rats, the expression of LINC00472 in the L4–L6 spinal cord dorsal tissue was elevated compared to rats in the sham operation group (control group) on post-operative days 0, 3, 7, 10, and 14 (Figure 1A). In addition, CCI rats were injected with a lentivirus vector containing LINC00472 (lv-LINC00472) and the transfection efficiency was verified (P<0.05; Figure 1B). The role of LINC00472 in the progression of neuropathic pain was assessed through measurements of the mechanical pain threshold and the cold pain threshold. Rats in the CCI group had significantly lower mechanical pain threshold and lower cold pain threshold in both feet compared to rats in the sham group at post-operative days 7, 10, and 14. In CCI rats treated with lv-LINC00472, both the mechanical pain threshold and the cold pain threshold were further reduced (Figure 1C-1F). These results demonstrated that upregulation of LINC00472 aggravated neuropathic pain in CCI rats, suggesting that LINC00472 is involved in the progression of neuropathic pain.
Downregulation of LINC00472 reduces inflammation
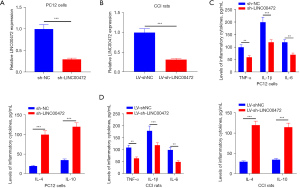
Previous studies have reported that inflammatory factors play a vital role in the progression of neuropathic pain (24). To determine the role of LINC00472 in the progression of neuropathic pain mediated by inflammatory factors, LINC00472 expression was suppressed in rat pheochromocytoma PC12 cells using short hairpin (sh)-LINC00472 and the corresponding negative control (sh-NC) (P<0.05; Figure 2A). LINC00472 expression was suppressed in the CCI rats by transfection of lv-sh-LINC00472 and the corresponding control, lv-shNC, into the spinal cord (P<0.05; Figure 2B). The levels of inflammatory biomarkers including TNF-α, IL-1β, IL-6, IL-4, and IL-10 in the cells and rat models were assessed. As shown in Figure 2C, TNF-α, IL-1β, and IL-6 levels were downregulated by sh-LINC00472, while IL-4 and IL-10 levels were upregulated (P<0.05; Figure 2C). Similarly, downregulation of LINC00472 reduced the levels of TNF-α, IL-1β, and IL-6 in the L4–L6 spinal cord of the CCI rats, and increased the levels of IL-4 and IL-10 (P<0.05; Figure 2D). The above results suggested that LINC00472 is involved in the progression of neuropathic pain by mediating the expression of inflammatory mediators.
LINC00472 targets the microRNA miR-300
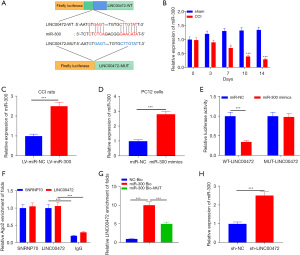
Bioinformatics analysis identified miR-300 as a potential target of LINC00472 in neuropathic pain. The binding between LINC00472 and miR-300 was examined using constructs containing the wild-type LINC00472 binding site (WT-LINC00472) and a mutant LINC00472 binding site (MUT-LINC00472) (Figure 3A). The expression of miR-300 in the L4–L6 spinal dorsal tissue of CCI rats was downregulated compared with control rats on days 0, 3, 7, 10, and 14 after the operation (Figure 3B). Further experiments were conducted in which lv-miR-300 was injected into CCI rats (Figure 3C) and miR-300 mimics were transfected into PC12 cells (Figure 3D). Co-transfection of miR-300 mimics and a luciferase vector containing WT-LINC00472 resulted in a significant decrease in relative luciferase activity in HEK-293T cells (Figure 3E). To determine whether LINC00472 could be used as the sponge RNA of miR-300, RIP analysis was performed. The results demonstrated that LINC00472 and miR-300 were more abundant in Ago2 particles than in IgG particles (Figure 3F). In addition, the RNA pull-down test using the biotinylated miR-300 (miR-300-bio) probe increased the level of LINC00472 compared to the miR-300 probe in the control group (NC-bio) (Figure 3G). Furthermore, low expression of LINC00472 elevated the levels of miR-300 in PC12 cells (Figure 3H). These results indicated that miR-300 is a direct microRNA target of LINC00472 and its expression is negatively regulated by LINC00472.
High mobility group box protein 1 (HMGB1) is a direct target of miR-300
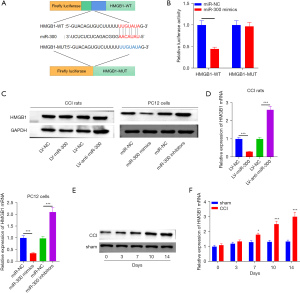
Bioinformatics analysis (http://www.targetscan.org/vert_72/) identified HMGB1 as a potential downstream target of miR-300. The binding site between miR-300 and HMGB1 is shown in Figure 4A. The luciferase reporter analysis was used to assess the relationship between miR-300 and HMGB1. As shown in Figure 4B, luciferase activity was significantly reduced in 293T cells co-transfected with miR-300 and HMGB1-WT. The protein and mRNA expression of HMGB1 decreased with miR-300 and lv-miR-300 treatment, but increased with miR-300 inhibitors and lv-anti-miR-300 treatment, suggesting that miR-300 negatively regulates HMGB1 expression in PC12 cells and CCI rats (Figure 4C,4D). In addition, the expression of HMGB1 in the spinal cord of CCI rats was also detected in a time-dependent manner. The protein and mRNA expression of HMGB1 was elevated in CCI rats compared to sham rats (Figure 4E,4F). These results suggested that miR-300 targets HMGB1 and negatively regulates its expression in PC12 cells and the CCI rat model, and indeed this may represent a potential mechanism by which miR-300 facilitates neuropathic pain.
LINC00472 regulated neuropathic pain progression through the miR-300/HMGB1 axis
To investigate whether LINC00472 regulates neuropathic pain through HMGB1, lv-sh LINC00472 and lv-anti-miR-300 were co-transfected into CCI rats. Rats with lv-sh LINC00472 showed reduced expression of HMGB1, while rats with lv-anti-miR-300 demonstrated elevated levels of HMGB1 (Figure 5A). Furthermore, rats with lv-sh LINC00472 showed significantly increased mechanical pain threshold and cold pain threshold, and this effect was reversed by lv-anti-miR-300 (P<0.05; Figure 5B-5E). Rats with lv-sh LINC00472 also showed decreased expression of IL-6, IL-1β, and TNF-α, and elevated expression of IL-4 and IL-10. Conversely, rats with lv-anti-miR-300 showed a partial reversal of the effects of lv-sh LINC00472 on inflammatory markers (P<0.05; Figure 5F,5G). This suggested that the interaction between LINC00472/miR-300/HMGB1 is related to neuropathic pain.
Discussion
Neuropathic pain accounts for about one-fifth of all patients with chronic pain. It has a high incidence and it causes significant damage to the patient’s physical and mental well-being (25). In recent years, the role of non-coding RNAs, such as lncRNAs and miRNAs, in the pathogenesis of neuropathic pain has attracted much attention. This study demonstrated that LINC00472 is positively correlated with the development of neuropathic pain. In the CCI rat model, the expression of LINC00472 was significantly elevated and was associated with significantly accelerated development of neuropathic pain. Our study also demonstrated that LINC00472 induces neuropathic pain by stimulating the expression of inflammatory factors. Furthermore, the data indicated that LINC00472 may induce the progression of neuropathic pain by regulating the miR-300/HMGB1 axis. Thus, the report provided novel insights into the molecular mechanism of neuropathic pain, and this information may contribute to the development of future clinical treatments.
Thousands of intergenic lncRNAs have been identified by whole-genome sequencing and have been shown to play vital roles in a variety of diseases. LINC00472 has been identified as a novel type of lncRNA, consisting of 3 exons, that plays a key role in the gene regulation of many diseases, including malignancies. For example, LINC00472 can inhibit the migration and invasion of lung adenocarcinoma cells through regulation of YBX1 (26). Similarly, LINC00472 can also inhibit the progression of hepatocellular carcinoma by regulating the LINC00472/miR-93-5p/PDCD4 axis (27). In addition, in acute liver injury, LINC00472 aggravates the LPS-induced inflammatory response by regulating the miR-373-3p/TRIM8 axis (15). Our current study demonstrated that the expression of LINC00472 was increased in CCI rats, and upregulation of LINC00472 could aggravate the development of neuropathic pain. Studies have reported that neuroinflammation is also closely related to chronic pain (24). Our results revealed that downregulation of LINC00472 inhibited neuropathic pain by reducing the expression of neuroinflammation-related molecules including IL-6, IL-1β, and TNF-α, and elevating the expression of IL-4 and IL-10.
In recent years, studies have shown that the mutual regulation between lncRNAs and miRNAs plays an essential role in the occurrence and development of neuropathic pain. Ma et al. showed in the CCI model that downregulating the lncRNA MALAT1 inhibits the development of neuropathic pain through regulation of the miR-129-5p/HMGB1 axis (28). In addition, deletion of SNHG4 (small nucleolar RNA host gene 4) inhibited neuroinflammation and neuropathic pain by targeting miR-423-5p (29). In the current study, bioinformatics analysis identified miR-300 as a likely target of LINC00472. Previous research has demonstrated that miR-300 can participate in a variety of diseases. For example, the expression of miR-300 is upregulated in gastric cancer cell lines, and high expression of miR-300 is associated with poor prognosis of gastric cancer (30). Conversely, in a rat model of cancer bone pain, miR-300 reduced pain by targeting HMGB1 (20). However, the precise role of miR-300 in neuropathic pain remains unclear. In this current study, we demonstrated that LINC00472 could regulate neuropathic pain by regulating miR-300. The expression of LINC00472 was negatively correlation with miR-300 and overexpression of miR-300 could partially reverse the effects of LINC00472 in promoting the progression of neuropathic pain.
HMGB1 is a late inflammatory factor that belongs to the alarmin family. Together with the pathogen related molecular pattern family (PMMP), HMGB1 forms the damage-related molecular pattern family (DAMPs), which plays an important role in the occurrence and development of inflammatory responses. Previous studies have reported that HMGB1 aggravates LPS-induced acute lung injury through the TLR4/NF-κB and PI3K/Akt/mTOR pathways (31). Studies have also shown that in tumor-infiltrating dendritic cells, the interaction between the TIM-3 and HMGB1 receptors can inhibit nucleic acid-mediated innate immune responses (32). In recent years, there has been much focus on the role of HMGB1 in neurological diseases. HMGB1 can induce astrocyte inflammation through the NF-κB pathway (33). Other studies using the CCI rat model demonstrated that miR-129-5p regulates the expression of HMGB1 to reduce neuropathic pain (34). Consistent with previous studies, this present report found that HMGB1 was elevated in the CCI rat model. Furthermore, miR-300 targeted HMGB1 and negatively regulated its expression. Further experiments confirmed that LINC00472 could regulate the expression of HMGB1 by absorbing miR-300, and participate in the progression of neuropathic pain by mediating the level of inflammation.
This investigation revealed the important role of the LINC00472/miR-300/HMGB1 axis in neuropathic pain. LINC00472 knockout increased miR-300 expression and reduced HMGB1 expression, alleviated neuropathic pain, and inhibited neuroinflammation in CCI rats. However, there were several limitations to this study. First, clinical samples should be included in future studies to explore the role of LINC00472 in patients with neuropathic pain. In addition, the role of other potential downstream miRNA targets related to LINC00472 should be further explored. Previous studies have shown that HMGB1 interacts with the TLR4/NF-κB and PI3K/Akt/mTOR pathways to aggravate LPS-induced acute lung injury (29). The ability of HMGB1 to regulate relevant signaling pathways and mediate neuroinflammatory responses in neuropathic pain warrants further investigation. In conclusion, this study demonstrated the role and molecular mechanisms of LINC00472 in neuropathic pain, and this may provide insights into potential therapeutic strategies for patients with neuropathic pain.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-2651
Data Sharing Statement: Available at https://dx.doi.org/10.21037/apm-21-2651
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-2651). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were approved by The Shenzhen People’s Hospital, The First Affiliated Hospital of Southern University of Science and Technology Ethics Committee, and is compliance with The First Affiliated Hospital of Southern University of Science and Technology institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- St John Smith E. Advances in understanding nociception and neuropathic pain. J Neurol 2018;265:231-8. [Crossref] [PubMed]
- Cohen SP, Mao J. Neuropathic pain: mechanisms and their clinical implications. BMJ 2014;348:f7656. [Crossref] [PubMed]
- Meacham K, Shepherd A, Mohapatra DP, et al. Neuropathic Pain: Central vs. Peripheral Mechanisms. Curr Pain Headache Rep 2017;21:28. [Crossref] [PubMed]
- Echeverry Stefania, Xiang Qun Shi, Yang Mu, et al. Spinal microglia are required for long-term maintenance of neuropathic pain. Pain 2017;158:1792-1801. [Crossref] [PubMed]
- Senba E, Kami K. Potentiation of spinal GABA inhibition as a therapeutic target for chronic neuropathic pain: from transplantation to physical exercise. Ann Palliat Med 2020;9:2430-6. [Crossref] [PubMed]
- Finnerup NB, Kuner R, Jensen TS. Neuropathic Pain: From Mechanisms to Treatment. Physiol Rev 2021;101:259-301. [Crossref] [PubMed]
- Jathar S, Kumar V, Srivastava J, et al. Technological Developments in lncRNA Biology. Adv Exp Med Biol 2017;1008:283-323. [Crossref] [PubMed]
- Wu Z, Ouyang C, Peng L. Risk assessment model and nomogram established by differentially expressed lncRNAs for early-stage lung squamous cell carcinoma. Transl Cancer Res 2020;9:5304-14. [Crossref]
- Yi M, Dai X, Li Q, et al. Downregulated lncRNA CRNDE contributes to the enhancement of nerve repair after traumatic brain injury in rats. Cell Cycle 2019;18:2332-43. [Crossref] [PubMed]
- Xia WQ, Niu GZ, Yin CG, et al. Effects of lncRNA gm4419 on rats with hypertensive cerebral atherosclerosis through NF-κB pathway. Eur Rev Med Pharmacol Sci 2019;23:10976-81. [PubMed]
- Liu Y, Feng L, Ren S, et al. Inhibition of lncRNA DILC attenuates neuropathic pain via the SOCS3/JAK2/STAT3 pathway. Biosci Rep 2020;40:BSR20194486 [Crossref] [PubMed]
- Tang W, Zhang L, Li Z. Long noncoding RNA LOC100911498 is a novel regulator of neuropathic pain in rats. Brain Behav 2021;11:e01966 [Crossref] [PubMed]
- Mao X, Zhou X, Liu J, et al. Up-regulated Linc00472 suppresses development of lung cancer cell via inhibition of MiR-196b-5p. Biosci Biotechnol Biochem 2019; Epub ahead of print. [Crossref] [PubMed]
- Wang Z, Katsaros D, Biglia N, et al. ERα upregulates the expression of long non-coding RNA LINC00472 which suppresses the phosphorylation of NF-κB in breast cancer. Breast Cancer Res Treat 2019;175:353-68. [Crossref] [PubMed]
- Li L, He Y, He XJ, et al. Down-regulation of long noncoding RNA LINC00472 alleviates sepsis-induced acute hepatic injury by regulating miR-373-3p/TRIM8 axis. Exp Mol Pathol 2020;117:104562 [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351-79. [Crossref] [PubMed]
- Marangon D, Raffaele S, Fumagalli M, et al. MicroRNAs change the games in central nervous system pharmacology. Biochem Pharmacol 2019;168:162-72. [Crossref] [PubMed]
- Zhang X, Zhang Y, Cai W, et al. MicroRNA-128-3p Alleviates Neuropathic Pain Through Targeting ZEB1. Neurosci Lett 2020;729:134946 [Crossref] [PubMed]
- Liu C, Yang J, Liu H, et al. miR-300 mitigates cancer-induced bone pain through targeting HMGB1 in rat models. Genes Genomics 2020;42:309-16. [Crossref] [PubMed]
- Xian S, Ding R, Li M, et al. LncRNA NEAT1/miR-128-3p/AQP4 axis regulating spinal cord injury-induced neuropathic pain progression. J Neuroimmunol 2021;351:577457 [Crossref] [PubMed]
- Wu J, Wang C, Ding H. LncRNA MALAT1 promotes neuropathic pain progression through the miR-154-5p/AQP9 axis in CCI rat models. Mol Med Rep 2020;21:291-303. [PubMed]
- Zhang J, Zhang J, Zhang D, et al. Down-regulation of LINC00472 promotes osteosarcoma tumorigenesis by reducing FOXO1 expressions via miR-300. Cancer Cell Int 2020;20:100. [Crossref] [PubMed]
- Hung AL, Lim M, Doshi TL. Targeting cytokines for treatment of neuropathic pain. Scand J Pain 2017;17:287-93. [Crossref] [PubMed]
- Gilron I, Baron R, Jensen T. Neuropathic pain: principles of diagnosis and treatment. Mayo Clin Proc 2015;90:532-45. [Crossref] [PubMed]
- Deng X, Xiong W, Jiang X, et al. LncRNA LINC00472 regulates cell stiffness and inhibits the migration and invasion of lung adenocarcinoma by binding to YBX1. Cell Death Dis 2020;11:945. [Crossref] [PubMed]
- Chen C, Zheng Q, Kang W, et al. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clin Res Hepatol Gastroenterol 2019;43:436-45. [Crossref] [PubMed]
- Ma X, Wang H, Song T, et al. lncRNA MALAT1 contributes to neuropathic pain development through regulating miR-129-5p/HMGB1 axis in a rat model of chronic constriction injury. Int J Neurosci 2020;130:1215-24. [Crossref] [PubMed]
- Pan X, Shen C, Huang Y, et al. Loss of SNHG4 Attenuated Spinal Nerve Ligation-Triggered Neuropathic Pain through Sponging miR-423-5p. Mediators Inflamm 2020;2020:2094948 [PubMed]
- Hong B, Li J, Huang C, et al. miR-300/FA2H affects gastric cancer cell proliferation and apoptosis. Open Med (Wars) 2020;15:882-9. [Crossref] [PubMed]
- Meng L, Li L, Lu S, et al. The protective effect of dexmedetomidine on LPS-induced acute lung injury through the HMGB1-mediated TLR4/NF-κB and PI3K/Akt/mTOR pathways. Mol Immunol 2018;94:7-17. [Crossref] [PubMed]
- Chiba S, Baghdadi M, Akiba H, et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol 2012;13:832-42. [Crossref] [PubMed]
- Al-Ofi EA, Al-Ghamdi BS. High-mobility group box 1, an endogenous ligand of toll-like receptors 2 and 4, induces astroglial inflammation via nuclear factor kappa B pathway. Folia Morphol (Warsz) 2019;78:10-6. [PubMed]
- Tian J, Song T, Wang W, et al. miR-129-5p Alleviates Neuropathic Pain Through Regulating HMGB1 Expression in CCI Rat Models. J Mol Neurosci 2020;70:84-93. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


