
Evaluation of minor drug-resistant viral variants in patients experiencing virological failure (VF) on a first-line regimen in Fujian Province by high-throughput sequencing
Introduction
The detection of drug resistance is an essential part of the clinical management of antiretroviral therapy for AIDS patients (1). However, commonly used in-house genotypic drug resistance detection methods based on Sanger sequencing fail to detect 20% of inferior drug-resistant strains (2,3). This affects the analysis of drug resistance in patients with virological failure (VF) and raises questions such as, how much additional burden does inferior drug-resistant mutation bring to the drug resistance of patients; does primary PIs class resistance mutation change before and after first-line treatment; and will it affect the therapeutic effect of second-line treatment with PIs? In this study, a high-throughput sequencing method established by Dawn M. Dudley. was conducted to redesign the sequencing primers suitable for epidemic strains in this region (4). Drug resistance mutations were detected in patients who failed to receive first-line treatment, and the results were compared with the traditional genotypic drug resistance detection results, to fully understand drug resistance gene variation.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/apm-21-1347).
Methods
Research object
Fifteen HIV patients who had received first-line antiviral therapy for more than one year and with a viral load more than 1,000 copies/mL were randomly selected for analysis. All HIV/AIDS cases were confirmed as HIV positive by Western blot (WB) test in the Fujian Provincial AIDS laboratory, and a corresponding epidemiological case investigation was conducted. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethical Review Committee of Fujian Municipal Center for Disease Control and Prevention and informed consent was taken from all the patients.
Main instruments and reagents
The following were used:
- Instrument: Roche/454 GS junior;
- Accessories: BDD (bed disposition device), suspended drop PCR sequencing bead counter, BDD centrifuge balance plate, and BDD centrifuge adapter;
- Kit: GS FLX titanium rapid library preparation kit, GS FLX titanium rapid library mid adapters kit, GS junior titanium emPCR oil and breaking kit, and GS junior titanium sequencing kit;
- Software: Amplicon Variant Analyzer (AVA).
Experimental methods
Amplification library preparation
RNA extraction: 140 µL plasma was taken and RNA was extracted by QIAamp RNA Blood Mini Kit in strict accordance with the instructions.
The first round of nested PCR: one step RT-PCR (see Table 1 for primer information).

Full table
PCR reaction conditions: 50 °C 60 min, 94 °C 2 min, 94 °C 15 sec, 60 °C 30 sec, 68 °C 30 sec ×2, 94 °C 15 sec, 58 °C 30 sec, 68 °C 30 sec ×2, 94 °C 15 sec, 55 °C 30 sec, 68 °C 30 sec ×2, and 68 °C 10 min.
The second round of nested PCR: the 2199–3285 region of pol gene was amplified by three fragments, and labeled with Pro, RT-A, and RT-B. The sequence information of the target region primer is shown in Table 2.
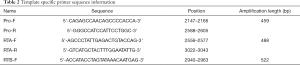
Full table
PCR reaction conditions: 94 °C 2 min, 94 °C 15 sec, 60 °C 30 sec, 68 °C 30 sec ×2, 94 °C 15 sec, 58 °C 30 sec, 68 °C 30 sec ×2, 94 °C 15 sec, 55 °C 30 sec, 68 °C 30 sec ×16, and 68 °C 10 min.
Purification, quality control, quantification, dilution, and enrichment of amplicon library.
emPCR and sequencing
emPCR and Roche 454 were sequenced with the qualified amplified library prepared by the above method.
Filtering of original sequencing data
The original sequencing data was split according to MID using Mothur software (version 1.31.2, http://www.mothur.org/), and all reads were assigned to the corresponding samples. The sequenced sequences were converted into FASTQ format. According to the mass fraction, three ends (average mass fraction at least 17) were pruned, and Roche/454 amplification adapters were removed in addition to the sequences shorter than 50 bp.
Alignment and SNP calling
Subsequent sequence alignment with HXB2 reference strain (GenBank NCA_001802). A bam alignment sequence was generated by BWASW, then GATK call snp was used with the parameter “stand_ call_ conf 50-stand_ emit_ conf 10”. The bases of all alignment sequences were then evaluated, and those with a mass fraction of more than 19 were called single nucleotide polymers (SNPs), while those with a mass fraction of more than 25 were called deletion or insertion polymorphisms. After processing, the SNPs of the relative reference strains were listed to obtain the number of sequences covering the mutation sites and the frequency of each mutation in the sample, and further drug resistance analysis was performed.
Drug resistance analysis
According to the known drug resistance mutations in the Stanford HIV drug resistance data base (Stanford DHIVDB), the specific amino acid changes at corresponding sites could be listed. The results of SNP calling were compared with them, and the proportion of sequences containing drug resistance mutations at a certain site in the total sequences covering that site were calculated. Drug resistance mutations accounting for less than 20% were defined as inferior drug resistance mutations.
Statistical analysis
A Chi square test was used to compare the mean numbers of DRMs detected by high-throughput versus standard sequencing in 14 samples for all mutations, nucleoside reverse transcriptase inhibitor (NRTI) mutations, NNRTI mutations and protease inhibitor (PI) mutations, respectively. Differences were considered statistically significant at a P value <0.05, all P values were calculated and reported in Table 3.

Full table
Results
Characteristics of drug resistance mutation
While 15 patients were enrolled in the study, valid data for analysis was obtained from only 14. The basic information of patients is shown in Table 4.
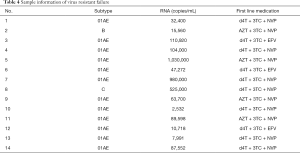
Full table
A total of 109 drug-resistant mutations were found in the 14 patients with valid sequencing data, of which low-frequency mutations accounted for 19.27% (21/109), with a lowest frequency of 1.3%. At least one NRTIs and NNRTIs high-frequency mutation was found in these samples (Table 5). Among NRTIs, M184V accounted for the highest proportion, reaching a staggering 100% (14/14), mainly because Lamivudine was included in the first-line regimen, and this mutation was selected by it. Of the thymidine analog mutations (TAMs) found, the main types were D67N (50%, 7/14), K219Q (50%, 7/14), T215F (42.86%, 6/14), and K70R (28.57%, 4/14). In NNRTIs, G190A was the most common mutation (64.29%, 9/14), followed by Y181C (42.86%, 6/14), and K103N (42.86%, 6/14). No high-frequency major drug-resistant mutation was found in PIs, and only one low-frequency major mutation I84V (1.69%, 29/1,709) was detected in 13 samples.
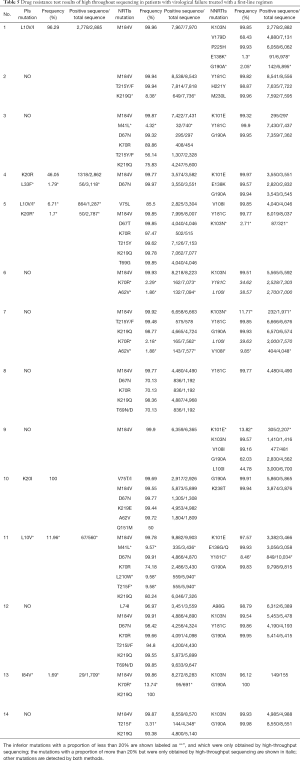
Full table
Compared with the conventional Sanger method, 24 additional mutations were detected in 14 samples, with an average of 1.7 mutations per sample, and the frequency of these mutations was less than 40%. Even if these mutations were divided into three categories, the differences remained statistically significant (Table 3).
Effect of drug-resistant mutation on drug resistance burden of patients with VF
We used the Stanford HIV drug resistance scoring algorithm (Stanford HIV drug resistance scoring algorithm: https://hivdb.stanford.edu/hivdb/by-mutations/) to measure the impact on the burden of drug resistance of patients caused by the additional inferior mutations found by high-throughput sequencing. Inferior drug resistance mutations were detected in 11 samples, and except for No. 4, the drug resistance score of the remaining 10 samples to at least two drugs increased, and the drug resistance grade of nine samples to at least one drug increased. For example, No. 7 added the inferior mutation A62V and K70R found by high-throughput sequencing, and the M184V, T215F and K219Q sites were detected by both methods. The grade of drug resistance (zidovudine, AZT) changed from moderate to high, and after adding the inferior resistance mutation, No. 11 showed that it was highly resistant to all NRTIs and NRTIs. The specific data are shown in the Table 6.
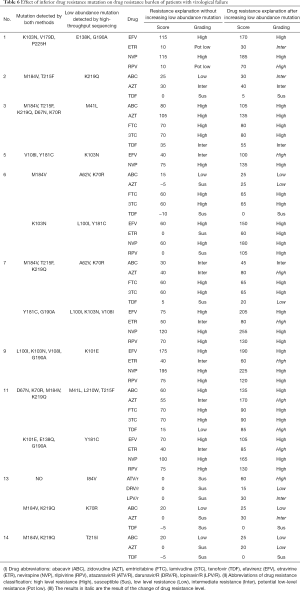
Full table
Screening of novel drug-resistant mutation sites
In view of the huge amount of sequence information that can be obtained by high-throughput sequencing, we attempted to determine whether there were novel mutations affecting drug resistance by comparing the sequence mutations of untreated and failed patients. Firstly, at least five mutations with a mutation rate higher than 90% were screened out from the sequencing results of treatment failure patients, and 56 base mutation sites were obtained. After eliminating the known drug resistance mutation, polymorphism mutation, and mutation outside the catalytic activity region, the remaining 14 sites were found. Chi square test was then used to compare the detection results of these mutation sites in the treatment group and untreated group one by one, and the results showed that there were three mutations in the two groups with significant difference (P≤0.01) and no literature report (see Table 7 for details). We then verified the phenotypic resistance of these three mutations, especially I135T.

Full table
Discussion
In addition to the high genetic diversity of HIV, the inferior drug-resistant mutation can also be the result of the spread of drug-resistant strains in infected individuals (5-7). This may not only lead to treatment failure but is often the manifestation of primary drug resistance. Therefore, the detection of inferior drug-resistant mutations is of great significance for monitoring the prevalence of primary drug-resistant strains, formulating appropriate initial treatment plans for AIDS patients, analyzing the causes of virological failure, and studying the mechanism of viral resistance.
Genotypic drug resistance testing has been carried out in patients with VF in Fujian Province since 2008, Yansheng Yan and other scholars (8-11) analyzed the drug resistance gene variation of HIV-1 strains in this and other provinces in China in recent years. The results showed that M184V and G190A were the most common sites of NRTI and NNRTI resistance, respectively, which are consistent with our results. All patients had at least one NRTIs and NNRTIs high-frequency mutation, and the highest proportion of NRTIs was M184V, followed by TAMs: D67N, K219Q, T215F, and K70R. G190A was the most common mutation in NNRTIs, followed by Y181C and K103N, and M184V + TAMs + G190A was the most common mutation combination. The high proportion of M184V raises concerns because 3TC has always been a first-line and second-line drug, raising the prospect that this protocol may need to be reconsidered.
Palmer (12) reported that 1–10 low-frequency drug resistance was detected by single gene sequencing from each patient with treatment failure. Our research shows that it is difficult to detect low-frequency mutations with conventional methods when virology fails. However, with a high-throughput sequencing method, 1.7 more mutations were detected in each sample on average, and even if these mutations were divided into three categories according to drug types, the difference is statistically significant. Low frequency mutations with frequency less than 20% account for 19.27% of all mutations, and we used the Stanford HIV resistance score system to demonstrate these mutations detected by high-throughput sequencing increased the burden of genotype resistance. Of the samples with inferior drug resistance mutations 90.9% (10/11) increased the drug resistance score of at least two drugs, and 81.8% (9/11) of the samples with inferior drug resistance mutations increased the drug resistance grade of at least one drug. For example, after adding the inferior mutation A62V and K70R found by high-throughput sequencing, plus the sites M184V, T215F, and K219Q detected by both methods, the resistance level of zidovudine (AZT) changed from moderate to high. Of note, adding the inferior resistance mutation to No.11 showed that it was highly resistant to all NRTIs and NRTIs. We used high-throughput sequencing to provide additional evidence that the burden of routine sequencing is much higher than that of routine methods.
The number of people receiving antiviral treatment in Fujian Province has increased from 100 in 2005 to more than 2000 in 2013. Most of these patients take AZT/d4T + 3TC + NVP/EFV as the first-line treatment, while PIs are left as second-line treatment. Previous studies have found that the resistance rate to PIs in AIDS patients with virological treatment failure is minimal (13), but as most of these studies used the traditional genotypic drug resistance test, it is impossible to determine whether there are inferior mutations of PI resistance below the detection threshold. If there were, the question of whether this affects the subsequent use of second-line treatment containing protease inhibitors is raised. Our experimental results showed that there were few drug-resistant mutations in the PI region in patients with VF which was consistent with previous reports, and there were few inferior drug-resistant mutations, with only one patient having the inferior major drug-resistant mutation I84V (1.69%, 29/1,709). It has been reported that the inferior mutants containing PI drug-resistant mutations are rarely detected in the untreated population, and that this will not significantly influence the effect of subsequent treatment regimens containing PI (14,15). Our results show inferior mutants containing PI drug-resistant mutations are rarely detected in patients with VF after treatment, and this suggests a more optimistic therapeutic effect for subsequent drug use when PIs are added as a second-line regimen.
We also attempted to determine whether there might be suspicious new mutation sites affecting drug resistance by comparing the sequence mutations of untreated and failed patients. As a result, three suspicious new mutation sites (I72V, I135T, I142S) were screened out and future research is required to investigate these sites. It is evident that in the screening of suspected drug resistance sites, a high-throughput sequencing method has considerable advantages. While the use of hundreds of thousands of sequences and tens of thousands of mutations make it possible to screen new sites, it is difficult to determine whether information processing means are available. Our analysis method represents a first attempt, and there is a need for more advanced data processing methods to make better use of the obtained high-throughput sequencing data.
In short, high-throughput sequencing methods have shown extremely high clinical and scientific value in the study of HIV inferior drug-resistant mutations. In the future, if the operation process can be improved and the average cost can be further reduced, it will certainly benefit more AIDS patients.
Acknowledgments
Funding: The study was sponsored by the Leading project of Fujian Science and Technology Department (2016Y0010).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-1347
Data Sharing Statement: Available at https://dx.doi.org/10.21037/apm-21-1347
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-1347). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethical Review Committee of Fujian Municipal Center for Disease Control and Prevention and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hirsch MS, Brun-Vézinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis 2003;37:113-28. [Crossref] [PubMed]
- Brun-Vézinet F, Costagliola D, Khaled MA, et al. Clinically validated genotype analysis: guiding principles and statistical concerns. Antivir Ther 2004;9:465-78. [PubMed]
- Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Top HIV Med 2008;16:266-85. [Crossref] [PubMed]
- Dudley DM, Chin EN, Bimber BN, et al. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One 2012;7:e36494 [Crossref] [PubMed]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 1995;267:483-9. [Crossref] [PubMed]
- Metzner KJ, Rauch P, Walter H, et al. Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS 2005;19:1819-25. [Crossref] [PubMed]
- Palmer S, Boltz V, Maldarelli F, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 2006;20:701-10. [Crossref] [PubMed]
- Yan Y, Wu S, Chen L, et al. Shift in HIV/AIDS Epidemic in Southeastern China: A Longitudinal Study from 1987 to 2015. Int J Environ Res Public Health 2016;13:794. [Crossref] [PubMed]
- Wu S, Qiu Y, Yan P, et al. Effectiveness of first-line antiretroviral therapy in HIV/AIDS patients: A 5-year longitudinal evaluation in Fujian Province, Southeast China. Arch Virol 2015;160:2693-701. [Crossref] [PubMed]
- Wang J, Wang Z, Liu J, et al. Efficacy and HIV drug resistance profile of second-line ART among patients having received long-term first-line regimens in rural China. Sci Rep 2015;5:14823. [Crossref] [PubMed]
- Liu H, Ma Y, Su Y, et al. Emerging trends of HIV drug resistance in Chinese HIV-infected patients receiving first-line highly active antiretroviral therapy: a systematic review and meta-analysis. Clin Infect Dis 2014;59:1495-502. [Crossref] [PubMed]
- Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 2005;43:406-13. [Crossref] [PubMed]
- Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naïve subjects in the CASTLE study. PLoS One 2010;5:e10952 [Crossref] [PubMed]
- Zhao T, Chen J, Fang H, et al. Diffusion tensor magnetic resonance imaging of white matter integrity in patients with HIV-associated neurocognitive disorders. Ann Transl Med 2020;8:1314. [Crossref] [PubMed]
- WHO UNICEF, UNAIDS. Towards universal access: scaling up priority HIV/AIDS interventions in the health sector. Progress Report 2010.
(English Language Editor: B. Draper)


