
Aggravation of airway inflammation in RSV-infected asthmatic mice following infection-induced alteration of gut microbiota
Introduction
Asthma is a chronic disease that affects children with variable expiratory airflow limitation. Some of its symptoms include wheezing, shortness of breath, chest tightness, and cough due to chronic airway inflammation (1). According to the Global Institute for Asthma (GINA) Committee and the International Study Asthma and Allergies in Childhood (ISAAC) Phase Three Study Group, 300 million people are affected with asthma worldwide, with 3.2% of cases in the Asia-Pacific region and 4.9% of children suffering from severe asthma worldwide (1,2). In China, the incidence, prevalence, and disability-adjusted life years (DALYs) of childhood asthma continue to rise (3,4).
Respiratory syncytial virus (RSV) is a negative-sense single-stranded RNA virus of the Paramyxoviridae family (5), and is the most common respiratory pathogen. RSV infection can promote airway obstruction and recurrent wheezing, leading to airway damage, which is a huge disease burden in infants and young children globally (6). Furthermore, asthma can also lead to more frequent and severe RSV infection (7). Asthma with RSV infection can release multiple mediators and cytokines that amplify the inflammatory response in asthmatic airway structural cells and participate in the structural changes in the airways. This leads to the induction of mucus hypersecretion, subepithelial fibrosis, and an increase in smooth muscles and blood vessels in the airway walls. Taken together, these changes result in airway inflammation, narrowing and airway hyper-responsiveness (AHR) (7-9).
The gut microbiota comprises bacteria that colonize the gastrointestinal tract (GIT) and represents 10 bacterial species in the human gut (10). Healthy gut flora can promote nutrient metabolism and mucosa growth, which is mainly related to the host’s overall health (11). Recent studies have found that a change in the quantity, type, and maturation of gut microbiota at 3 months of age is a crucial factor affecting the development of asthma and atopy in childhood (11-13). Notably, symptoms of digestive system diseases, such as vomiting, diarrhea, and abdominal pain have been observed in patients with corona virus disease 2019 (COVID-19), the ongoing pandemic with a mortality rate of 1.4% (14). These findings are consistent with the ‘gut-lung axis’ theory, which has linked changes in the gut microbiome with lung inflammation, as lung infection can disrupt the gut microbiota via the blood circulatory system (15). Although it is known from the literature that RSV infection in asthma patients impacts their gut microbiota, the imbalances in the types of their gut microbiota and the mechanism of airway inflammation regulation in RSV-infected asthma patients is still unclear.
In this study, we created an RSV-infected OVA challenged mice and studied an RSV-infected asthma mouse model. We subsequently tested the mice lung function, lung histopathology, and the expression of inflammation cytokines (IL-5, IL-13, IL-25, and IL-33) using ELISA and 16S rRNA sequencing. Finally, we analyzed the relationship between immunoglobulin E (IgE), inflammatory cytokines, and gut microbiota.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/apm-20-2052).
Methods
Preparation of RSV
RSV was prepared according to the method described by Yan Sun (16). The RSV A2 strain (ATCC, USA) was inoculated into human laryngeal carcinoma cells (Hep-2 cells), Shanghai Cell Bank, Chinese Academy of Sciences (CAS). After 3–5 days, RSV A2 caused the Hep-2 cells to fuse and form a “multinucleated giant cell” syncytium. For complete and detailed steps of this process, please refer to our previous work (17). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013)
RSV infected-asthmatic mice
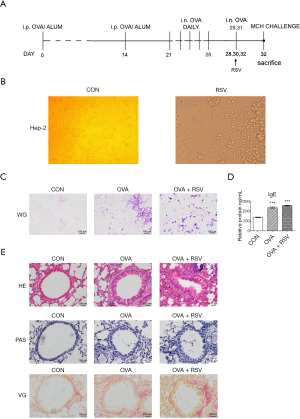
Six-to-eight-week-old BALB/c male mice (Shanghai SLAC Laboratory Animal Co., Ltd.) were provided with a standard diet and maintained at 24±2 °C, 46%±5% humidity in a light/dark cycle. After 5 days of acclimatization, 24 mice were randomly divided into a control group (CON, n=8), an ovalbumin group (OVA, n=8), and an OVA + RSV group (n=8). The asthma model was sensitized via intraperitoneal injections (i.p.) of OVA (grade V, Sigma-Aldrich, USA) (OVA 20 µg + AL [OH]3 2 mg, 0.2 mL) at 0, 14, and 28 days. The OVA and OVA + RSV groups received consecutive RSV infections (1.0×106 PFU·mL−1, 50 µL) in the morning of days 28, 30, and 32, while the mice from the control group were administered phosphate buffer saline (PBS) (Figure 1A). For complete and detailed steps of this process, please refer to our previous work (17). After the last administration, all mice were anesthetized with sodium pentobarbitone (2.5%). The bronchoalveolar lavage (BAL) fluid was collected for Wright-Giemsa stain, lung function was tested, and lung tissues were collected for examining inflammatory changes using haematoxylin and eosin (HE), periodic acid-Schiff (PAS), Van Gieson (VG) staining. The serum levels of interleukins (ILs) were analyzed using ELISA. Samples from the large intestines and feces were collected for 16S rRNA sequencing. Experiments were performed under a project license (No. 2016001) granted by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University, in compliance with the National Research Council criteria or institutional guidelines for the care and use of animals.
BAL fluid
After lung function assessment, the mice were sacrificed via administration of sodium pentobarbitone (2.5%) and cannulation of the trachea. Next, BAL obtained from the airway lamina was washed with 1.0 mL sterile PBS. Cell types were determined, and 100 mL fluid was deposited onto glass slides using cytospin for Wright-Giemsa staining. Differential cell counts were determined from 200–600 leukocytes using standard hematological criteria. The BAL fluid supernatant was stored at −25 °C.
Lung function of RSV infected-asthmatic mice
AHR was measured with Buxco’s modular and invasive system (Buxco Electronics Inc., NY, USA). Pulmonary airway resistance (RL) and dynamic lung compliance (Cdyn) were tested directly (as described previously) (17). The anesthetized mice were tracheostomized and intubated with a special cannula and then laid in the supine position inside a plethysmograph chamber. After a stable baseline airway pressure was achieved, the mice were successively administered aerosolized PBS. Subsequently, the values were recorded in response to increasing methacholine concentrations (3.125, 6.25, and 12.5 mg/mL) and were expressed as a percentage change from the baseline value.
Histology
Lung tissues from the mouse model were fixed in 4% buffered paraformaldehyde overnight, embedded in paraffin, and cut (Leica, Germany, RM2235) into 4 µm sections. HE, PAS, and VG staining were used to observe discrepancies in the lung tissue. The sections were then viewed under a light microscope using high magnification (400×) (Leica, Germany, BX42).
ELISA
The mice’s blood was collected after 2 h at 20–25 °C and then centrifuged for 30 min at 1,200 ×g/4 °C to isolate the serum. The levels of inflammatory factors (IgE, IL-5, IL-13, IL-25, and IL-33) were analyzed using ELISA according to the manufacturer’s instructions (USCN Business Co., Ltd., China).
16S rRNA sequencing
Isolated genomic DNA from the fecal samples of RSV-infected mice was polymerase chain reaction (PCR) amplified for the V3-V4 hypervariable regions of the bacterial 16S rRNA. The reaction was carried out in a 25 µL volume using universal primer pairs (343F: 5'-TACGGRAGGCAGCAG-3'; 798R: 5'-AGGGTATCTAATCCT-3'). Both primers were connected with an Illumina sequencing adapter. The reaction conditions were as follow: 94 °C for 5 min; 26 cycles at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 5 min.
The amplicons were purified with AMPure XP beads (Beckman Coulter, A63881, USA), visualized using gel electrophoresis, and amplified for the second round of PCR. Following amplicon purification using the AMPure XP beads, the final amplicon was quantified by Qubit dsDNA assay kit (Life Technologies, Q32854, USA). Equal quantities of purified amplicon were pooled for sequencing. Two paired-end read cycles of 300 bases each to sequence were performed on an Illumina Miseq (Illumina Inc., USA).
Bioinformatics
To detect and cut off ambiguous bases (N), paired-end reads were preprocessed using Trimmomatic v 0.35 software (18). Reads with a low quality score (below 20) were cut off using the trimming approach. Next, paired-end reads were assembled using the Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) v1.2.11 software (19). The assembly parameters were as follows: 10 bp of minimal overlapping, 200 bp of maximum overlapping, and 20% of maximum mismatch rate. Further denoising of the sequencing data was performed as follows: reads with ambiguous, homologous sequences, or those below 200 bp, were not considered. Seventy-five percent of bases above Q20 were retained using Quantitative Insights into Microbial Ecology (QIIME) 1.8.0 software (http://qiime.org/) (20). The reads with chimera were subsequently detected and removed using UCHIME v 4.2.40 (21). Clean reads were subjected to primer sequence removal and clustering to generate operational taxonomic units (OTUs) using VSEARCH software (version 10.0.240, http://www.drive5.com/usearch) with a 97% similarity cut off (22). Each OTU was selected using the QIIME package (http://qiime.org/). All representative reads were annotated and blasted against the Silva database (Version 123) using a Ribosomal Database Project (RDP) classifier v 2.2 (confidence threshold was 70%) (23). The microbial diversity of fecal samples in RSV-infected mice was estimated using the alpha diversity, including the Chao1 (24) and Shannon indexes (25). The Bray Curtis distance matrix was generated using QIIME software, which also provides the Bray Curtis Principal Coordinates Analysis (PCoA) and phylogenetic tree (26) construction. The 16S rRNA sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China).
Data availability
All bacterial sequencing data of 16S rRNA genes generated in this study were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accession number PRJNA627257 (http://www.ncbi.nlm.nih.gov/sra).
Statistical analysis
All data were statistically analyzed and presented as mean ± standard deviation (SD) using GraphPad Inc., San Diego, CA, USA (GraphPad Prism V5, USA). The three groups were analyzed using one-way analysis of variance (ANOVA), and multiple comparisons were analyzed using t-tests. ELISA comparisons between two groups were analyzed using one-way ANOVA. A two-sided P<0.05 was considered statistically significant.
Results
RSV-inoculated and RSV-infected asthmatic mice
After RSV-inoculated Hep-2 cells were infected with the RSV A2 strain, typical fusion lesions (syncytia formations) could be viewed under the microscope. This was fused into a ‘multinucleated giant cell’ syncytium
One of the important features of the allergic asthma model is the production of Ovalbumin-specific IgE upon induction. So, levels of IgE were measured in the serum of the control (137.57±4.92), OVA (236.45±11.34), and OVA + RSV (257.37±6.79) groups. IgE levels in the serum of OVA challenged mice were significantly higher than in the control mice (P<0.05), and OVA + RSV group mice had significantly higher IgE levels than the OVA challenged group (P<0.01) (Figure 1D).
To define the inflammatory changes in the established RSV-infected asthma model, pathological manifestation of the lung was evaluated by H&E staining. Compared with the control group mice, an obvious presence of inflammatory cells around the airways and vessels was observed in the OVA challenged group. The number of inflammatory cells presents around the airways of asthmatic mice infected with RSV dramatically increased compared with the other groups (Figure 1E).
Our team also studied goblet cell hyperplasia and mucus hyperproduction by PAS staining and lung peribronchial collagen deposition using VG staining. The results showed that RSV-infected asthmatic mice developed significantly aggravated goblet cell hyperplasia and mucus hyperproduction and increased peribronchial collagen deposition than the other groups (Figure 1E).
Aggravation of AHR and IL cytokine expression in RSV-infected asthmatic mice
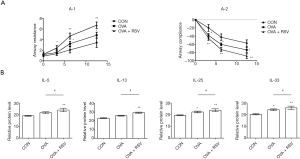
The RSV-induced aggravation of AHR in the OVA-induced asthma model was also evaluated. We found that mild changes in RL and Cdyn were observed in the control group mice. However, there was a significant change in airway responsiveness in the OVA-exposed and OVA + RSV group mice, with an obvious increase in RL and decreased Cdyn compared with the control mice. The OVA + RSV group exhibited significantly aggravated airway responsiveness provoked by methacholine and decreased airway compliance compared with the OVA and control groups (P<0.05, P<0.01) (Figure 2A). RSV-infected asthmatic mice were found to increase the expression of IL cytokines, such as IL-5 (CON 19.01±1.49, OVA 21.66±2.24, OVA + RSV 23.95±4.33); IL-13 (CON 23.03±1.31, OVA 25.80±1.22, OVA + RSV 29.24±1.78); IL-25 (CON 19.44±0.89, OVA 22.32±1.78, OVA + RSV 23.83±2.60); and IL-33 (CON 20.23±1.53, OVA 24.07±2.00, OVA + RSV 25.80±4.30). RSV-infected asthmatic mice could give rise to abnormal production of cytokines. As shown in Figure 2B, the serum IL levels were higher in the OVA group and highest in the OVA + RSV group, compared with the control group (P<0.05 and P<0.01, respectively).
The composition of the gut microbiome is altered after RSV-infection in asthmatic mice
The feces of mice were analyzed to investigate whether RSV-infection affects the gut microbiota of asthmatic mice. The gut microbiota was visualized using a flower plot and community bar plot created using the sequencing data. The flower plot suggested that asthma and RSV-infected asthma significantly altered the number of OTUs. There were 157 core OTUs in the three groups, and the number of OTUs in the OVA + RSV group was significantly reduced (Figure 3A). From the community bar plot, it was possible to observe that there were differences among the three groups at the levels of phylum (Figure 3B), family (Figure 3C), and genus (Figure 3D).
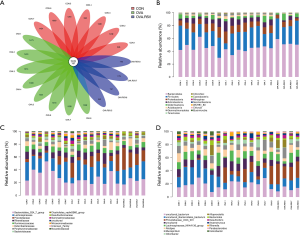
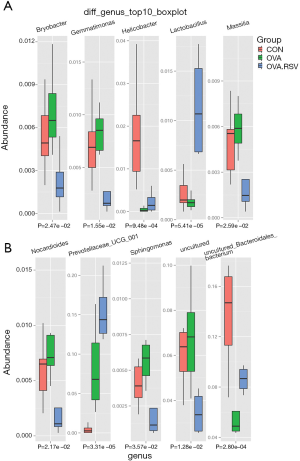
Compared with the CON and OVA groups, the abundance of Bacteroidetes and Firmicutes were higher and lower, respectively, in the OVA + RSV group. In this result, the eight DNA samples extracted (CON groups) did not have good quality; thus, only seven samples were analyzed. From the OVA + RSV groups 1–4, the DNA extracted samples did not have good quality, so we could only analyze 5–8 samples after eliminating the 1–4 data sets. In order to verify the quality of the reserved samples, we verified them twice. Compared with the CON group, the abundance of both Bacteroidetes and Firmicutes were lower in the OVA group, which may be due to the lower abundance of Bacteroidales_S24_7_group, Bacteroidaceae, Clostridiales_vad in BB60_group, and Lachnospiraceae. Significant differences were observed in the abundance of Prevotellaceae_UCG_001 and Helicobacter. Compared with the control group, the abundance of both Bacteroidetes and Firmicutes were higher in the RSV group, which may be due to the growth of Prevotellaceae, Porphyromonadaceae, Bacteroidaceae, and Lachnospiraceae. The significant abundance differences were verified for Lachnospiraceae_NK4A136_group, Prevotellaceae_UCG_001, Alloprevotella, Desulfovibrio, Mucispirillum, and Alistipes. Compared with the OVA group, the abundance of both Bacteroidetes and Firmicutes were higher in the OVA + RSV group, which may be due to the growth of Bacteroidales_S24_7_group, Prevotellaceae, and Lachnospiraceae. Significant differences were verified for Prevotellaceae_UCG_001, Lachnospiraceae_NK4A136_group, Alloprevotella, Desulfovibrio, Mucispirillum, and Alistipes.
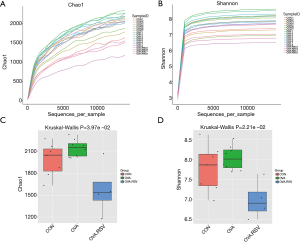
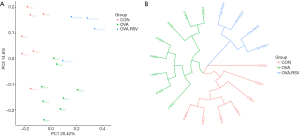
The alpha exponential rarefaction curve of the Chao1 index (Figure 4A) and the Shannon index (Figure 4B) showed clear asymptotes, which indicated a near-complete sampling of the community. The box plots of the Chao1 index (Figure 4C) and the Shannon index (Figure 4D) exhibited significant differences among the three groups. Beta diversity statistics using the Principal Coordinate Analysis (PCoA) revealed significant differences in the cluster patterns of gut microbial communities (Figure 5A,B).
Changes in the gut microbiota in RSV infected asthmatic mice
Multivariate statistical analysis (ANOVA) screened the top 10 genera with significant differences and high relative abundance: Bryobacter, Gemmatimonas, Helicobacter, Lactobacillus, Massilia, Nocardioides, Prevoteliaceae_UCG_001, Sphingomonas, and two uncultured bacteria (Figure 6A,B).
To identify the key biomarkers that are differentially displayed between the three groups, we performed a statistical analysis using linear discriminant analysis effect size (LEfSe) analysis with linear discriminant analysis (LDA) to characterize different species between the groups.
We found that the taxonomic levels from kingdom to genus corresponded to the circles radiating from the inside out (Figure 7A). In the cladogram, the red, green, and blue nodes represented the microbial taxa that play an important role in the CON, OVA, and OVA + RSV groups, respectively.
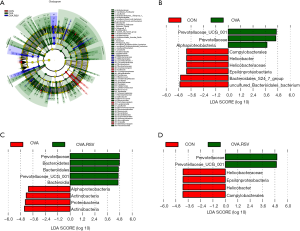
In the LDA scores histogram, biomarkers with significant differences between groups were revealed, and the score values represented the influence degree of the corresponding taxa. As shown in Figure 7B,C,D, there were 9, 9, and 6 differential bacterial taxa in CON, OVA, and OVA + RSV group, respectively, and 15 microbial taxa, namely Prevotellaceae_UCG_001, Prevotellaceae, Alphaproteobacteria, Campylobacterales, Helicobacter, Helicobacteraceae, Epsilonproteobacteria, Bacteroidales_S24_7_group, Bacteroidetes, Bacteroidales, Bacteroidia, Actinbacteria, Proteobacteria, and Uncultured_Bacteroidales_bacterium. Prevotellaceae_UCG_001, Helicobacter, and Bacteroidales_S24_7_group could serve as potential diagnostic and treatment biomarkers among the groups.
Alteration of gut microbiota was associated with the levels of IgE and ILs in RSV-infected asthmatic mice
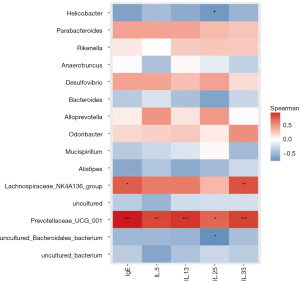
To further verify the potential function of these core OTUs, we examined the reliability relationship between the core OTUs and IgE and cytokines (including IL-5, IL-13, IL-25, and IL-33), and found that 15 microbial OTUs were significantly associated with the IgE and inflammatory cytokines. Prevotellaceae_UCG_001, which is positive, was the most significantly associated with IgE, IL-13, and IL-33 (P<0.001), and was even more significantly associated with IL-5 (P<0.01) and IL-25 (P<0.05). Lachnospiraceae_NK4A136_group is also positive, and was significantly associated with IgE (P<0.05) and IL-33 (P<0.01). Helicobacter and Uncultured_Bacteroidales_ bacterium, which are negative, have relevance and consistency with IL-25 (P<0.05) (Figure 8).
Discussion
Globally, asthma affects the health of an increasing number of people, which is expected to rise to 400 million by 2025. Approximately 250,000 asthma-related deaths are reported annually (26). RSV-related asthma is associated with worse aggravation, higher risk, intensive care, and mortality in some cases (27). Some studies have shown that RSV significantly alters gut microbiota diversity (28). In this study, an increase in Bacteroidetes and a decrease in Firmicutes phyla abundance have been observed in RSV-infected mice.
Gut microbiota plays an important role in nutrient metabolism, immunomodulation, drug metabolism, and xenobiotic/antimicrobial protection (11). Bacteroidetes and Firmicutes comprise two major phyla of the normal human gut microbiota. Recent research suggests that an imbalance of the abundance and diversity in gut microbiota has a compact connection with airway microbiota and asthma alterations, especially in children (29,30). It is believed that the GIT and respiratory tract share the same mucosal immune system, which is one of the mechanisms of the gut-lung axis (31). In this study, we focused on gut microbiota changes in RSV-infected asthma mice and characterized the abundance and diversity changes in the gut microbiota following RSV infection.
We discovered that OVA-challenged asthma led to a decrease in both the phylum of Bacteroidetes and Firmicutes. Both phyla exhibited an intense decrease after RSV infection, which even exceeded the normal group. According to the family and OTU levels, Prevotellaceae_UCG_001, Lachnospiraceae_NK4A136_group, Alloprevotella, Desulfovibrio, Mucispirillum, and Alistipes brought out the growth of Bacteroidales_S24_7_group, Prevotellaceae, and Lachnospiraceae, which supported a Bacteroidetes and Firmicutes blast. The unknown regulation of RSV resulting in Bacteroidetes and Firmicutes propagation is noteworthy and constitutes a part of our next study.
RSV is commonly responsible for acute respiratory disease in children and adults, and RSV-infected asthmatic mice showed an enhanced serum level of IgE (32,33). Th2 cytokines, such as IL-5 and IL-13, play an important role in RSV-aggravated asthma (34-36). The high level of IL-5 recruits and activates eosinophil proliferation. An increase in IL-13 level induces AHR. RSV-infected asthma can also provoke epithelium-derived cytokines, such as IL-25, IL-33, leading to inflammation, which facilitates asthma development (37). IL-33 enhances AHR and airway inflammation by suppressing antiviral responses (38). Previous research suggested that viral infection induces airway hyper-reactivity via the IL-13-IL-33 axis pathway (39).
Gut microbiota impacts cytokines and fecal microbiota transplantation in gut microbiota-depleted model mice leads to a normalization of pulmonary bacterial counts (40). In this study, we performed a correlation analysis, which showed that ILs positively correlate with RSV infection with different major genus. Our results showed that Prevotellaceae_UCG_001, which is positive, was most significantly associated with IgE, IL-13, IL-33 (P<0.001), and even more significantly with IL-5 and IL-25. Lachnospiraceae_NK4A136_group, which is also positive, was significantly associated with IgE and IL-33 (P<0.01). Helicobacter and Uncultured_Bacteroidales_bacterium, which are negative, had relevance and consistency with IL-25 (P<0.05). Prevotellaceae_UCG_001 and Lachnospiraceae_NK4A136_group especially expressed a significant correlation with ILs, which suggested potential therapeutic targets for RSV-infected asthma following alteration of gut microbiota. Meanwhile, Prevotellaceae_UCG_001 and Lachnospiraceae_NK4A136_group were selected as potential therapeutic targets, as they presented significant differences following RSV infection.
Some studies in humans and mice found that the immune responses that improve and ameliorate disease caused by viral infections have used various Lactobacilli as probiotics (41). It has been proposed that enriching the gut microbiota with Lactobacillus could protect against airway inflammation in RSV infection (42). This study found that a few gut microbiota in RSV-infected asthmatic mice, such as Prevotellaceae_UCG_001 and Lachnospiraceae_NK4A136_group, could play an important role in airway inflammation caused by RSV-infected asthmatic mice.
Although it enriches the lung-gut axis theory and provides some new clues, our research has certain limitations that should be noted. Since RSV-infected asthma in children is associated with a dysbiosis of the gut microbiota, which is marked by a general decline in beneficial potential microbiota, it can enrich potentially pathogenic taxa. These changes in the gut microbiota might disrupt the gut barrier integrity and enhance gut-lung inflammation. In future research, we will further explore the changes in the gut microbiota of RSV-infected asthma patients.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant No. 81674020 to X Jing). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the arrive reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-2052
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-2052
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-2052). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. 2016001) granted by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University, in compliance with the National Research Council criteria or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global Strategy for Asthma Management and Prevention. Global Initiative for Asthma, 2020. Available online: www.ginasthma.org
- Lai CK, Beasley R, Crane J, et al. Global variation in the prevalence and severity of asthma symptoms: phase three of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2009;64:476-83. [Crossref] [PubMed]
- Mattiuzzi C, Lippi G. Worldwide asthma epidemiology: insights from the Global Health Data Exchange database. Int Forum Allergy Rhinol 2020;10:75-80. [Crossref] [PubMed]
- Chinese Tebo. Journal of Pediatrics respiratory group.Pediatric Society of Chinese medicine, children’s asthma Guidelines for diagnosis and treatment (2016 Edition). Chinese J Pediatr 2016;54:167-81.
- Nyiro JU, Munywoki P, Kamau E, et al. Surveillance of respiratory viruses in the outpatient setting in rural coastal Kenya: baseline epidemiological observations. Wellcome Open Res 2018;3:89. [Crossref] [PubMed]
- Rezaee F, Linfield DT, Harford TJ, et al. Ongoing developments in RSV prophylaxis: a clinician's analysis. Curr Opin Virol 2017;24:70-8. [Crossref] [PubMed]
- Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol 2017;140:895-906. [Crossref] [PubMed]
- Piedimonte G. Contribution of neuroimmune mechanisms to airway inflammation and remodeling during and after respiratory syncytial virus infection. Pediatr Infect Dis J 2003;22:S66-74; discussion S74-5. [Crossref] [PubMed]
- Tan YR, Yang T, Liu SP, et al. Pulmonary peptidergic innervation remodeling and development of airway hyperresponsiveness induced by RSV persistent infection. Peptides 2008;29:47-56. [Crossref] [PubMed]
- Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635-8. [Crossref] [PubMed]
- Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastroenterol 2015;21:8787-803. [Crossref] [PubMed]
- Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015;7:307ra152 [Crossref] [PubMed]
- Stokholm J, Blaser MJ, Thorsen J, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018;9:141. [Crossref] [PubMed]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020;382:1708-20. [Crossref] [PubMed]
- Sze MA, Tsuruta M, Yang SWJ, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS one 2014;9:e111228 [Crossref] [PubMed]
- Sun Y, López CB. Preparation of Respiratory Syncytial Virus with High or Low Content of Defective Viral Particles and Their Purification from Viral Stocks. Bio Protoc 2016;6:e1820 [Crossref] [PubMed]
- Jing X, Yan W, Zeng H, et al. Qingfei oral liquid alleviates airway hyperresponsiveness and mucus hypersecretion via TRPV1 signaling in RSV-infected asthmatic mice. Biomed Pharmacother 2020;128:110340 [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Reyon D, Tsai SQ, Khayter C, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012;30:460-5. [Crossref] [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016;4:e2584 [Crossref] [PubMed]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261-7. [Crossref] [PubMed]
- Chao A, Bunge J. Estimating the Number of Species in a Stochastic Abundance Model. Biometrics 2002;58:531-9. [Crossref] [PubMed]
- Hill TC, Walsh KA, Harris JA, et al. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 2003;43:1-11. [Crossref] [PubMed]
- Christiansen SC, Zuraw BL. Treatment of Hypertension in Patients with Asthma. N Engl J Med 2019;381:1046-57. [Crossref] [PubMed]
- Ritchie AI, Farne HA, Singanayagam A, et al. Pathogenesis of Viral Infection in Exacerbations of Airway Disease. Ann Am Thorac Soc 2015;12:S115-32. [PubMed]
- Rangelova E, Wefer A, Persson S, et al. Surgery Improves Survival After Neoadjuvant Therapy for Borderline and Locally Advanced Pancreatic Cancer: A Single Institution Experience. Ann Surg 2021;273:579-86. [Crossref] [PubMed]
- Lee-Sarwar KA, Kelly RS, Lasky-Su J, et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol 2019;144:442-54. [Crossref] [PubMed]
- Sharma A, Laxman B, Naureckas ET, et al. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol 2019;144:1214-27.e7. [Crossref] [PubMed]
- Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol 2017;15:55-63. [Crossref] [PubMed]
- Wang J, Wu J, Kong L, et al. BuShenYiQi Formula strengthens Th1 response and suppresses Th2-Th17 responses in RSV-induced asthma exacerbated mice. J Ethnopharmacol 2014;154:131-47. [Crossref] [PubMed]
- Xing QQ, Liu LW, Zhao X, et al. Serum proteomics analysis based on label-free revealed the protective effect of Chinese herbal formula Gu-Ben-Fang-Xiao. Biomed Pharmacother 2019;119:109390 [Crossref] [PubMed]
- Yoon HK, Shim YS, Kim PH, et al. The TLR7 agonist imiquimod selectively inhibits IL-4-induced IgE production by suppressing IgG1/IgE class switching and germline ε transcription through the induction of BCL6 expression in B cells. Cell Immunol 2019;338:1-8. [Crossref] [PubMed]
- Ito R, Maruoka S, Soda K, et al. A humanized mouse model to study asthmatic airway inflammation via the human IL-33/IL-13 axis. JCI Insight 2018;3:e121580 [Crossref] [PubMed]
- Ikutani M, Ogawa S, Yanagibashi T, et al. Elimination of eosinophils using anti-IL-5 receptor alpha antibodies effectively suppresses IL-33-mediated pulmonary arterial hypertrophy. Immunobiology 2018;223:486-92. [Crossref] [PubMed]
- Gauvreau GM, O'Byrne PM, Boulet LP, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med 2014;370:2102-10. [Crossref] [PubMed]
- Ravanetti L, Dijkhuis A, Dekker T, et al. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J Allergy Clin Immunol 2019;143:1355-70.e16. [Crossref] [PubMed]
- Chang YJ, Kim HY, Albacker LA, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol 2011;12:631-8. [Crossref] [PubMed]
- Schuijt TJ, Lankelma JM, Scicluna BP, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016;65:575-83. [Crossref] [PubMed]
- Zelaya H, Alvarez S, Kitazawa H, et al. Respiratory antiviral immunity and immunobiotics: beneficial effects on inflammation-coagulation interaction during influenza virus infection. Front Immunol 2016;7:633. [Crossref] [PubMed]
- Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A 2014;111:805-10. [Crossref] [PubMed]


