
The pharmacological mechanism of Huashi Baidu Formula for the treatment of COVID-19 by combined network pharmacology and molecular docking
Introduction
The 2019 new coronavirus [severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] epidemic was first reported in Wuhan, China, in December 2019, and has been declared by the World Health Organization as an urgent public health event of international concern, which evolved into a large-scale epidemic-related pandemic (1). Coronavirus disease 2019 (COVID-19) is caused by SARS-CoV-2, a virus that belongs to the coronavirus of the genus β. The virus has a diameter of 60–140 nm, and the particles are round or oval; the shape is often polymorphic. SARS-CoV-2 cell invasion depends on TMPRSS2, ACE2, and other main proteins, and its invasion mechanism is similar to that of SARS-CoV (2,3). After the virus invades, the body dysregulates inflammatory factors, resulting in excessive inflammation, which can accelerate the disease progress and produce clinical manifestations such as multiple organ failure (4). Patients with mild disease have symptoms such as fever, fatigue, and dry cough, and severe cases may have dyspnea, respiratory distress syndrome, or septic shock, and there are currently no specific antiviral drugs available in the clinic (1). As of Nov 29, 2020, more than 62.34 million people have been diagnosed with COVID-19, and the number of deaths is caused by the disease is close to 1,460,000 (5).
When the immune system is overactivated due to infection, disease, drugs, and other factors, immune cells will produce large amounts of cytokines, intensifying the inflammation. Many secreted pro-inflammatory factors will cause a positive feedback loop to break through a certain threshold, eventually forming a cytokine storm (6). Patients infected with SARS-CoV-2 with severe COVID-19 have a significant increase in pro-inflammatory cytokines such as IL-6, IL-7, TNF-α, IFN-γ, and their symptoms are characterized by a cytokine storm (7-9). The body produces inflammatory factors after the virus invades, causing an inflammatory overreaction, accelerating the disease and clinical manifestations such as single or multiple organ failure (10,11). In addition to the antiviral treatment and supportive care, it is necessary to monitor for cytokine storm in patients, as early treatment positively affects the reduction of critical conversion rate and reduces the mortality rate (12). In the treatment of COVID-19, Traditional Chinese medicine (TCM) and humanized monoclonal antibodies play a significant role in reducing the cytokine storm syndrome in COVID-19, including tocilizumab, Lianhuaqingwen, other antibodies targeting inflammatory cytokines, and other TCM prescriptions (13-15).
TCM has achieved good clinical results in preventing and treating new infectious diseases, especially in the prevention and treatment of COVID-19 (16-18). “Three Chinese Patent Medicines, Three Traditional Chinese Medicine Compounds” has been proven effective in the treatment of COVID-19. Huashi Baidu Formula (HSBDF) has been recommended for treating epidemic toxin blocking in the lung, a pattern seen in severe cases of COVID-19. HSBDF consists of 14 herbs, including Ephedra Herba, Amygdalus Communis Vas, Gypsum Fibrosum, licorice, Pogostemon Cablin (Blanco) Benth., Magnolia Officinalis Rehd Et Wils, Atractylodes Lancea (Thunb.)Dc., Amomum Tsao-Ko Crevostet, Pinelliae rhizoma preparata, Poria Cocos(Schw.) Wolf., Radix Rhei Et Rhizome, Hedysarum Multijugum Maxim., Lepidii Semen Descurainiae Semen, and Radix Paeoniae Rubra (19).
In 75 severe cases treated in Wuhan Jinyintan Hospital, the CT diagnosis showed significant improvement after using HSBDF in lung inflammation and clinical symptoms, and nucleic acid-negative time conversion and hospital stay time were shortened by an average of 3 days (20). In another study, including 124 moderate cases in Street Health Center at Jiangjun Road and 894 mild and moderate cases (452 in the TCM group) in Dongxihu Makeshift Cabin Hospital, showed that there were significant differences in the time to achieve symptoms relief and nucleic acid negative conversion after using HSBDF (21).
Although numerous studies have been carried out on the clinical efficacy, not many studies exist which initially reveal the potential mechanism HSBDF use may have on COVID-19, and the pharmacological mechanism remains unclear. Network pharmacology is a novel technology, and it improves the efficiency of drug discovery based on the component-target-pathway multi-network level. It has been widely used to screen active components, the mechanism of drug action, and the pathogenesis of diseases (22,23). It would be an essential tool for researching complex systems of TCM with the continuous accumulation of disease and drug-related targets, and the constant mining of chemical components of TCM (24). Molecular docking is a theoretical simulation method to predict its binding mode and affinity based on the characteristics of the receptor and the interaction between the receptor and drug molecules (25). This study aims to investigate the potential therapeutic mechanism of HSBDF on COVID-19 through network pharmacology combined with molecular docking. The flow chart of the research is shown in Figure 1.

We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/apm-20-1759).
Methods
The Component Database establishment
The components of HSBDF were initially searched in the TCMSP (http://tcmspw.com/tcmsp.php) and YaTCM (http://cadd.pharmacy.nankai.edu.cn/yatcm/home) databases. Then, using PubChem (https://pubchem.ncbi.nlm.nih.gov/), the SMILES structures were acquired, resulting in the Mol2 structure of all compositions, establishing the component database (26,27). The TCMSP and YaTCM databases are TCM databases with information on various herbs, chemicals, and targets, which are beneficial to study TCM scientifically and precisely, and the PubChem database is the central database of chemical research (28).
The component database was established according to the compendial flow chart (Figure 2). We initially collected the herbal medicine information, including 14 crude drug medicinal materials. The TCMSP and YaTCM databases were used to obtain the component information of the 14 crude drug medicinal materials. Pharmacokinetic and drug screening models were used to screen the active ingredients from the chemical compositions, including the Obioavail 1.1 system, the Tanimoto coefficient, and Lipinski Rule of Five, and the parameters included DL ≥0.18, OB ≥30%, Lipinski Rule of Five ≥0.75 (29-32). The T(a,b) index was defined as follows:

a represents the descriptor vector of the tested compound, and b represents the database descriptor vector of drug or drug-like compound.
After collecting the active ingredients, the chemical structural formula (the Mol2 2D structure, the Mol2 3D structure, the SMILES structure) was obtained from the TCMSP, YaTCM, and PubChem databases, as well as the properties of the active ingredients.
The active components acquirement and ADME screening
The absorption, distribution, metabolism, and excretion (ADME) screening, including the oral bioavailability (OB) and drug-like (DL) properties, was performed in all components to acquire active components from crude drug medicinal materials. The condition parameters were OB ≥30% and DL ≥0.18, Lipinski Rule of Five ≥0.75, which effectively screened the active components. The information was normalized using the settings of the OBioavail 1.1 system and the Tanimoto coefficient, which are efficient in collecting the active components (29,31).
Potential targets prediction and crucial targets screening
The prediction of the potential target was carried out using the TargetNet database (http://targetnet.scbdd.com/) for netting or predicting potential targets. It constructs QSAR models based on current chemogenomics data to make future predictions (33). We included models with AUC ≥0.7, and the result score of ≥0.5 was used for precise and efficient prediction. After gaining the active targets of HSBDF, disease targets of COVID-19 were obtained from other reports, which identified 5,555 genes co-expressed with ACE2 (34), which has been identified as the crucial cellular receptor for SARS-CoV and SARS-CoV-2. Then we used a Venn diagram to merge and intersect component target genes and COVID-19 disease genes to retrieve crucial target genes of HSBDF on SARS-CoV-2. Finally, the essential target genes database was established, and the genes were annotated and normalized using the Uniprot database (https://www.uniprot.org/) (35).
Protein-Protein Interaction (PPI) network construction
The PPI network was constructed through the STRING database (https://string-db.org/cgi/input.pl), a database of protein-protein interactions that covers 24,584,628 proteins from 5,090 organisms (36-37). We then visualized the data with the Cytoscape software (38). The Molecular Complex Detection (MCODE) plug-in for critical targets analysis was used with the following setting parameters: Degree Cutoff: 2, Node Score Cutoff: 0.2, K-Core: 2, and Max Depth: 100 (39).
The Gene Ontology (GO) analysis
The key targets were analyzed using the Metascape (http://metascape.org/gp/index.html) and OmicsBean (http://www.omicsbean.cn/) databases (40). Metascape uses the Benjamini-Hochberg P value correction algorithm and well-adopted hypergeometric test. In Metascape and OmicsBean databases, the investigation was based on fundamental and routine parameters that had a statistically significant difference (P value <0.05). Moreover, Cytoscape and other graphic software were used to draw related charts based on the GO analysis (41).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
A KEGG pathway enrichment was carried out using the Metascape database to find essential targets involved in physiological regulation (42). Differential expression gene and protein lists were used to design the KEGG analysis experiment, to allow for the detection of essential target proteins enriched in associated biological pathways.
Network construction
Networks were constructed using the Cytoscape software, including a component-target (C-T) network, component-target-pathway (C-T-P) network, herb-target (H-T) network, target-pathway (T-P) network, and meridian-tropism (M-T) network. A topology analysis was performed in each network, including information of the node, edge, degree, betweenness, and closeness centrality, which aids in elucidating the nature of the networks (38).
Molecular docking
Molecular docking was executed using the COVID-19 Docking Server (https://ncov.schanglab.org.cn/index.php), which aims to test the correlation between active compositions and essential proteins related to COVID-19. The molecules were acquired by topology analysis from the C-T-P network, and the target proteins that are essential for the treatment of COVID-19 are listed in Tables 3,4 (43). There were three groups of drugs used for the molecular docking: a group of HSBDF compounds, a control group of chemical drugs, and a negative control group. We included five molecules for the group of HSBDF compounds: baicalein, ellagic acid, genkwanin, isotrifoliol, and rhein. We included six compounds for the control group of chemical drugs: Lopinavir, Ritonavir, Remdesivir, darunavir, Arbidol, and Chloroquine, which might have a potential effect on COVID-19 (44). Water was used to verify the correctness of the database in the negative control group. The D3Targets-2019-nCoV database (https://www.d3pharma.com/D3Targets-2019-nCoV/index.php) was also used to verify the binding energy between bioactive components and other main proteins related to SARS-CoV-2; detailed information is shown in the results table (45).
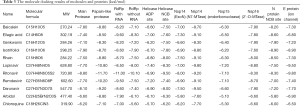
Full table
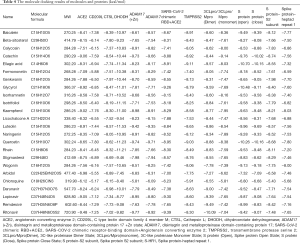
Full table
The study conformed to the provision of the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The quantitative data were analyzed with Student’s t-test using SPSS (25.0 version, SPSS Inc., Chicago, IL, USA). P value <0.05 was considered a statistically significant value.
Results
Identification compositions of HSBDF
The composition of HSBDF was acquired from the TCMSP and YaTCM databases, which included 223 bioactive compositions, and the basic information is listed in Table 1 and Figure 3. All medicinal chemistry information is listed in the supplementary information table (https://cdn.amegroups.cn/static/public/atm-20-1759-1.xlsx). We calculated the average of each chemical information (MW: 374.61, AlogP: 3.15, Hdon: 3.86, Hacc: 5.95, OB: 45.37%, Caco-2: 0.08, BBB: −0.50, DL: 0.49, FASA−: 0.29, HL: 10.48). Various compositions included glycoside compounds, ketones, steroid acid compounds, and other compounds rich in herbs, representing the diversity of compositions, revealing multi-compositions of HSBDF. Among those compositions, isoflavones and cynotoxin had a high OB score; (−)-medicocarpin and 9,10-dimethoxypterocarpan-3-O-β-D-glucoside had a high level of DL score.

Full table

Crucial targets of HSBDF on COVID-19
The 358 potential target genes were acquired using the TargetNet database, and the disease target genes related to ACE2 included 5,555 target genes built from the attachment in published reports. Finally, 84 crossed target genes were obtained (Figure 4), representing potentially crucial target genes for HSBDF on SARS-CoV-2. The essential target genes database was established after the annotation and normalization of the Unpriot database (Table 2), and most proteins found in the intersection targets were related to metabolism, sex hormones, and immunological stress.

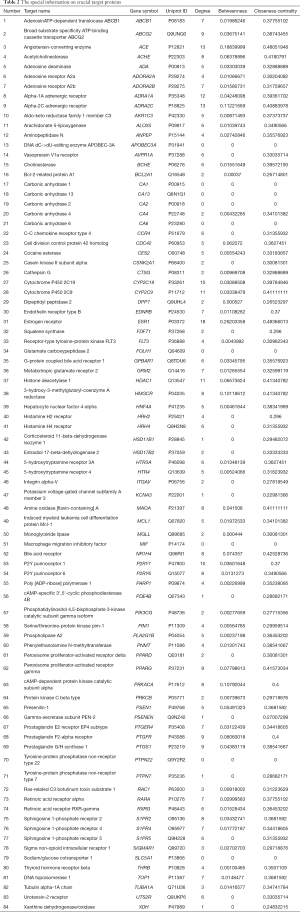
Full table
PPI network of crucial target proteins
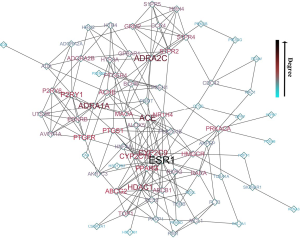
The 84 crucial target proteins were analyzed in the STRING database, and the PPI network was constructed using Cytoscape software (Figure 5). The network included 75 nodes, 211 edges, and 5,550 shortest paths. The essential proteins in the PPI network were ESR1, ADRA1A, ADRA2C, HDAC1, and ACE, which were considered the vital proteins for the treatment of HSBDF on COVID-19. The network was divided into seven submodules by the MCODE analysis (Figure 6), which revealed the particular interactions.

The GO biological analysis
The GO results were acquired after 84 essential proteins were analyzed by the Metascape and OmicsBean databases, and the overall histogram of GO biological enrichment was obtained using the OmicsBean database (Figure 7). For the biological processes, significant roles included the cellular response to the stimulus, the multicellular organismal process, which indicates the blocking of SARS-CoV-2 and the diversity action of the HSBDF components. The considerable role of cell components included the cytoplasm and cytosol, which could potentially be the therapeutic mechanism of HSBDF on COVID-19 since it might target the synthetic metabolism, primarily related to the protein, fat, and carbohydrate metabolism. For molecular functions, a significant role was found for protein binding, reflecting the mechanisms related to proteins consistent with the biological processes and the cell components involved. The specific bubble diagram and gene weight diagram are shown in Figure 8.

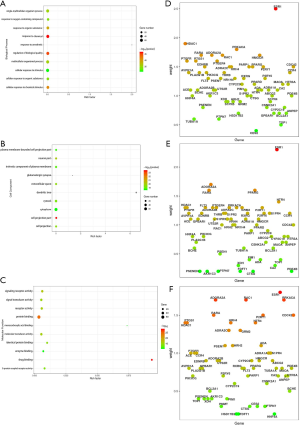
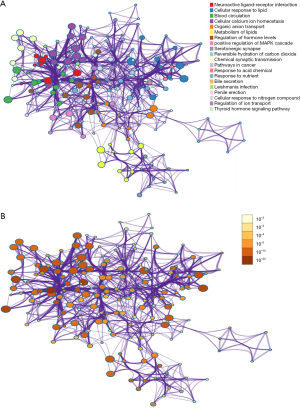
Based on the Metascape database, the interaction network of the GO biological process was acquired (Figure 9), which reflected the relationship of specific GO processes. Two graphs revealed the ties of GO biological enrichment and partial KEGG pathways, describing the interaction of biological processes. The significant role of the network included blood circulation, cellular calcium ion homeostasis, positive regulation of hormone levels, and positive regulation of the MAPK signaling cascade.
The KEGG pathway enrichment analysis
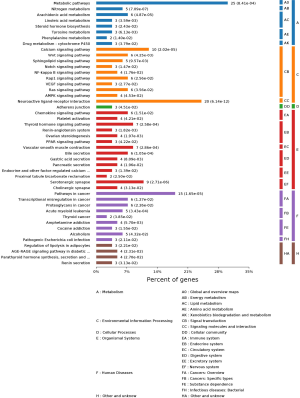
We found 46 pathways that were enriched in the KEGG pathway analysis performed using the OmicBean database. In the histogram of the KEGG pathways (Figure 10), vital pathways found included metabolic, calcium signaling, neuroactive ligand-receptor interaction, and cancer pathways. Crucial targets might be commonly enriched in the cancer-associated pathways since they are associated with an abundance of proteins, as we found in our previous study. The PPAR, AMPK, and NF-kappa B signaling pathways, renin-angiotensin system, renin secretion, vascular smooth muscle contraction, bile secretion, gastric acid secretion, and pancreatic secretion were also found to be vital pathways. Related graphs of the KEGG pathway analysis were also acquired, which elaborated on the therapeutic mechanism that HSBDF may have on COVID-19 from the perspective of the molecular pathway mechanisms (Figure 11). The mechanism diagram of HSBDF on SARS-CoV-2 is shown in Figure 12. And the more information about PPI, GO, and KEGG in Figures S1-S5.


Molecular docking
The 12 molecules were docked with 12 essential proteins by the COVID-19 docking server database, and the result is shown in Tables 3,4. It is generally believed that when the ligand's conformational stability and the receptor are stable, the lower the energy, the greater the possibility of binding, and most drugs in the control group had low binding energy. Meanwhile, some of the molecules also had low binding energy, for instance, ellagic acid with RdRp with RNA (−9.60 kcal/mol) and Nsp14 (−9.10 kcal/mol), isotrifoliol with N protein NCB site (−8.90 kcal/mol) and E protein (−8.90 kcal/mol), and rhein with Nsp14 (−9.10 kcal/mol). The molecular docking partially revealed the therapeutic mechanism of HSBDF on COVID-19. The visualization of docking results, including the 2D structure, is shown in Figure 13. The molecular docking results of the D3Targets-2019-nCoV database are shown in Table 4, it includes bioactive components and other main proteins directly related to SARS-CoV-2.

Network construction
The constructed networks are shown in Figure 14. The C-T-P network (Figure 14A) reflects the relationship between components, targets, and pathways. It includes 355 nodes, 1,575 edges, and 125,670 shortest paths, suggesting the crucial pathways in the use of HSBDF to affect SARS-CoV-2. The C-T network (Figure 14B) included 298 nodes, 1,575 edges, and 62,250 shortest paths, showing the relationship between components and targets without pathways and explains the mechanism from different perspectives. The H-T network (Figure 14C) describes the targets of 14 parts, which is beneficial for studying the treatment mechanism, and 93 nodes, 517 edges, and 8,556 shortest paths were involved in this network. The T-P network (Figure 14D) showed the relationship between targets and pathways. We found that PRKCB, PRKACA, and CDC42 are crucial targets, and the pathways associated with cancer, metabolic pathways, and neuroactive ligand-receptor interactions are essential pathways for the T-P network. Moreover, the T-P network included 125 nodes, 248 edges, and 15,550 shortest paths. The meridian-tropism network (Figure 14E) was significant for the TCM study, we found that the lung meridian, the spleen meridian, and the stomach meridian were potential target sites of HSBDF for the treatment of COVID-19, and there were 20 nodes, 38 edges, and 380 shortest paths covered in this meridian-tropism (M-T) network.

Discussion
TCM plays a significant role in the treatment of COVID-19, as most of the more than 50,000 discharged patients in China were treated with TCM as of Mar 15. The proportion of TCM applications in Wuhan city, Hubei province, and China was 89.10%, 91.64%, and 92.41%, respectively (19,21,46,47). HSBDF was gradually implemented in the treatment process of the first batch of Chinese medicine medical teams in Jinyintan Hospital and Dongxihu square cabin hospital (21,48). The use of HSBDF for the treatment of COVID-19 was based on multiple components, targets, and pathways that might significantly shorten the duration of the nucleic acid transfer, the average time of hospitalization, improve clinical symptoms, and improve findings at the physical and chemical examination and lung CT improvement (21,48). The experimental results showed that the viral load of lung tissue could be reduced by 30%, and in mice infected with SARS-CoV-2, the inflammation of the lung can be improved significantly with this drug (20).
Evidence-based medicine and systematic reviews show that the combination of TCM and standard care can significantly improve the treatment outcome in COVID-19 cases, including the overall treatment efficacy, the rate of symptom disappearance, and the improvement of other clinical symptoms (20,49). TCM has been found to have a positive effect on symptoms in a study among 732 adults in seven study groups, and no significant adverse events were recorded in the analysis (50). Recent evidence suggests that the remarkable effect of the combination of TCM and standard care reveals the clinical effect of Chinese medicine in the treatment of COVID-19 in a study including 855 patients in seven study groups (51). Existing research recognizes the critical role TCM played in treating COVID-19 in a study including 2,275 enrolled patients in 18 study groups, and improvements in clinical parameters, such as clinical cure rate, lung CT, inflammatory biomarkers, and other parameters were found (52). Recently, investigators have examined the effects of TCM on COVID-19 in a meta-analysis including 1,474 patients in 19 study groups. Combining Chinese and Western medicine to treat COVID-19 may effectively control symptoms and reduce disease progression, which is seen reflected by the improvement of symptoms, reduction of hospital stay, and conversion rate of critical illness of TCM (53). These findings have significant implications for the understanding of the clinical effect of HSBDF on COVID-19.
In this study, the mechanism behind the efficacy of HSBDF for the treatment of COVID-19 was studied based on network pharmacology and molecular docking. Databases and networks were applied to gain the herbs, components, targets, pathways, and the relationships between them. Using network pharmacology and molecular docking tools, the therapeutic mechanism of HSBDF on COVID-19 was preliminarily explained.
Natural products and TCM with high safety, high convenience, low toxicity, and broad-spectrum properties are widely used to prevent and treat infectious diseases, especially in the SARS and SARS-CoV-2 outbreaks (17,54). Many Chinese herbal extracts have antiviral abilities, as well as anti-inflammatory and immune regulatory functions. Some of the 223 biological components in HSBDF have such effects; for example, wogonin, baicalein, and baicalin have antiviral activity (55). Genkwanin can inhibit proinflammatory transmission to prevent the cytokine storm caused by SARS-CoV-2 infection via targeting the miR-101/MKP-1/MAPK pathway (56). Albiflorin may inhibit the inflammatory response through the LOX-1/NF-κB pathway (57). Ellagic acid has anti-inflammatory activity and inhibits HIV-1 infection in vitro, and has been shown to have antiviral activity against the Ebola virus, Human Papillomavirus (HPV), and human rhinoviruses (58-61). By the suppression of TLR/NF-κB and TLR/MAPK signaling pathways, isotrifoliol can inhibit pro-inflammatory mediators in vitro, which might inhibit the formation of a cytokine storm caused by SARS-CoV-2 infection (62). Besides, rhein attenuates inflammation by inhibiting NF-κB and NALP3 inflammasome, both in vivo and in vitro (63). For 223 components of HSBDF, the compositions play a significant direct role in the antiviral capability, the regulation of metabolism and synthesis, as well as the inflammatory immune regulation.
The mechanism of action of HSBDF on COVID-19 includes seven KEGG pathway components: (I) vascular function regulation pathways; (II) pathways associated with the immune response and inflammation; (III) metabolism pathways; (IV) sex hormone pathways; (V) microbial infection pathways; (VI) nerve-related pathways; and (VII) other pathways. The specific pathways mechanisms were analyzed as follows:
The vascular function regulation pathway includes vascular smooth muscle contraction, the VEGF signaling pathway, the renin-angiotensin system, and renin secretion. Its mechanism might be based on the indirect regulation of ACE2 both in the renin-angiotensin system and renin secretion (Figure 12), and HSBDF influences the regulation of renin and the biotransformation of angiotensin (64). The genetic deletion of ACE2 induces vascular dysfunction in mice, suggesting a relationship between ACE2 and vascular function, reflecting the association of COVID-19 and vascular function (65-68). Moreover, COVID-19 is closely related to cardiovascular diseases, and the regulation of cardiovascular diseases is beneficial for treatment, especially for critically ill patients (68,69). Because of the association of ACE2 with cardiovascular diseases, there is a potential for treatment with HSBDF by targeting the vascular function regulation pathway.
For pathways associated with the immune response and inflammation, we found significant roles for arachidonic acid metabolism, platelet activation, NF-kappa B signaling, chemokine signaling, and PPAR signaling (70-75). Notably, arachidonic acid is essential for prostaglandin, and is crucial for the prevention of a cytokine storm and is also the crucial target for cytochrome P450 (73). Therefore, arachidonic acid metabolism has the pharmacological effect of preventing a cytokine storm. For platelet activation, when the virus is invading the body, inflammation occurs. The immune defense mechanism is based on recruiting patrolling monocytes, and this recruitment process is based on the CCN1 protein generated from platelets and vascular endothelium, thus reflecting the significance of platelet activation (74). Moreover, through the NF-kappa B signaling pathway, HSBDF directly actives PKCβ protein and indirectly affects a series of complex proteins, affecting survival and inflammation. Through the immune response and inflammation pathways, HSBDF plays a pharmacological role in the treatment of COVID-19 by stoping the “cytokine storm”.
According to the single-cell RNA sequencing research of patient tissues, the digestive system is a vital target organ for SARS-CoV-2 infection, and metabolism and endocrine regulation is closely related to COVID-19 (76). Therefore, bile secretion, metabolic pathways, the AMPK signaling pathway, and other metabolic pathways are related to the treatment. The metabolism pathways intervene in the endocrinology and metabolism processes to protect the body, which reduces the number of cases from mild to severe symptoms, especially for patients with diabetes, cardiovascular disease, and metabolic disease. Due to the target organ infected by the virus, impairment of the intestinal barrier function would lead to the translocation of enteric bacteria and endotoxin, thus aggravating respiratory diseases of distal organs through intestinal lymphatic circulation. HSBDF could improve a patients' loss of appetite after infection with SARS-CoV-2, avoid deterioration of the disease, and affect the synthesis of cholesterol and protein to speed up the healing process (77).
A gender difference was found for infections with SARS-CoV-2, and the incidence and mortality in men are higher than that in women. A total of 41,510 out of 77,932 patients (53.3%) were males, and the mortality rates of male and female patients were 2.8% and 1.7%, respectively (78). Research elucidated the biological factors involved apart from differences in living habits: firstly, ACE2 is commonly expressed in prostate and testicles; secondly, ACE2 is positively correlated with the expression of androgen receptor (AR) and TMPRSS2, which is the crucial molecule that promotes the development of male prostate cancer and can be positively regulated by androgens in the body; thirdly, CCL14, CCL23, IL7, IL16, and IL1 which are pro-inflammatory cytokines and chemokines are also preferentially expressed in males, and the chemokines CCL2, CCL3, and CCL4 that could play a protective role are higher expressed in females; besides, T cells in the lungs of men were found to be less virus-killing than those found in women (2,78). For the effect of HSBDF on COVID-19, the regulation of sex hormone pathways consists of ovarian steroidogenesis and steroid hormone biosynthesis, and the virus infection process is intervened by the regulation of steroid hormone pathways, especially the sex steroid hormones.
Critically ill patients have neurological symptoms, which are manifested as acute cerebrovascular disease, unconsciousness, and skeletal myopathy (79), and HSBDF can be used for those patients. Moreover, through the neuropathic pathway, including neuroactive ligand-receptor interaction, serotonergic synapse, and the sphingolipid signaling pathway, HSBDF could potentially protect critically ill patients from neurological symptoms. Besides, some other pathways, such as pathogenic Escherichia coli infection, elaborate on the treatment mechanism of HSBDF on SARS-CoV-2. The antiviral and antimicrobial effects of HSBDF could be revealed from the view of microbial infections.
The therapeutic mechanism of HSBDF on SARS-CoV-2 can further be elucidated by molecular docking. We found that ellagic acid could dock well with RdRp and with RNA, in which the Nsp12 part has the function of RNA polymerase and plays a vital role in the replication and transcription of viral genomes (80). Ellagic acid could also dock well with Nsp14 (N7-MTase), which has a double enzyme activity, forms a complex with Nsp10 to function, and is responsible for capping mRNA (81). Isotrifoliol has a great docking ability with the N protein NCB site of Helicase (Nsp13), the ideal target for wide-spectrum antiviral drugs (82). Isotrifoliol could dock well with E protein, which forms pentapolymers and has ion channel functions, and HSBDF could cause interference with virus-related ion channels and block the toxic expression of the virus (83). Rhein could dock well with Nsp14 (ExoN), proofreading RNA replication and transcription, which reveals HSBDF might prevent the replication and transcription of RNA. Furthermore, the bioactive components of HSBDF also have an excellent binding ability with the proteins directly related to SARS-CoV-2. The discussion above explains the docking status between molecules of HSBDF and crucial proteins and indicates the therapeutic mechanism behind the effects of HSBDF for the treatment of COVID-19, which is mainly based on multiple effects of TCM.
While preliminary, this finding suggests that the pharmacological mechanism of HSBDF on COVID-19 is based on direct and indirect antiviral effects. The binding activity of active ingredients and crucial proteins related to SARS-CoV-2 plays direct antiviral effects, and the key biological pathway mechanisms of HSBDF on the body play indirect antiviral effects. This study has identified the active ingredients, such as isotrifoliol and ellagic acid, which have an excellent binding ability with crucial proteins directly related to COVID-19, including ACE2, ADAM17, and 3CLpro. Thus, it achieves a pharmacological effect through the combination of molecules and then directly achieves the therapeutic effect. Likewise, the indirect pharmacological mechanisms of HSBDF on COVID-19 are based on biological pathways, especially the crucial biological signaling pathways, including the AMPK signaling pathway, arachidonic acid metabolism, NF-kappa B signaling pathway, serotonergic synapse, renin secretion, and the renin-angiotensin system. The indirect pharmacological effects of HSBDF on COVID-19 are based on multiple target proteins of HSBDF on the body, such as AMPK, PPAR γ, arachidonic acid metabolism-related proteins, 5-HT3, and ACE, thus having mainly indirect pharmacological effects on metabolism, inflammation, and nerves. The contribution of this study has been to preliminarily confirm the potential pharmacological mechanisms of HSBDF on COVID-19, which provides a theoretical basis and a reasonable preliminary explanation for the clinical effects, including severe conversion (hospitalization) rate, the increase in total effective treatment rate, and the improvement in lung imaging signs and major related symptoms (50).
Above all, TCM plays an essential part in the antiviral process by targeting multiple components, targets, and pathways based on the network pharmacology and molecular docking, and different networks reflect the various properties of the mechanisms. Moreover, this research needs to be further improved and analyzed through experiments, and the molecular mechanism of HSBDF on COVID-19 should be deepened. Western-Blot (WB) and Polymerase Chain Reaction (PCR) experiments could be used for mechanism verification.
Conclusions
In summary, 223 components, 84 crucial target proteins, and 46 potential pathways were acquired by network pharmacology, molecular docking, and TCM theory, which preliminarily elaborated the therapeutic mechanism of HSBDF on COVID-19 at the molecular level. HSBDF may affect SARS-CoV-2 infection through multiple components, targets, and pathways.
Acknowledgments
Funding: The authors were supported by grants from the Project of the Ministry of Science and Technology of China (SQ2017YFC170086-3) and Research Project of Hubei University of Chinese Medicine (2020ZZX003).
Footnote
Reporting Checklist: The authors have completed Materials Design Analysis Reporting (MDAR) checklists. Available at http://dx.doi.org/10.21037/apm-20-1759
Peer Review File: Available at http://dx.doi.org/10.21037/apm-20-1759
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-1759). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020;323:1061-9. [Crossref] [PubMed]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181:271-80.e8. [Crossref] [PubMed]
- Xu J, Zhao S, Teng T, Guo X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020;12:244. [Crossref] [PubMed]
- Sanders JM, Monogue ML, Cutrell JB. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020;323:1824-36. [PubMed]
- Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020;20:533-4. [Crossref] [PubMed]
- Tisoncik JR, Korth MJ, Katze MG, et al. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 2012;76:16-32. [Crossref] [PubMed]
- Huang C, Wang Y, Cao B, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Grazioli S, Tavaglione F, Blanchard-Rohner G, et al. Immunological assessment of pediatric multisystem inflammatory syndrome related to COVID-19. J Pediatric Infect Dis Soc 2020;12:piaa142.
- Mehta P, McAuley DF, Brown MHLH Across Speciality Collaboration, et al. UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 2020;395:1033-4. [Crossref] [PubMed]
- Dzobo K, Chiririwa H, Dandara C, et al. Coronavirus Disease-2019 Treatment Strategies Targeting Interleukin-6 Signaling and Herbal Medicine. OMICS 2021;25:13-22. [Crossref] [PubMed]
- Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest 2020;130:2202-5. [Crossref] [PubMed]
- Henderson LA, Canna SW, Nigrovic PA, et al. On the Alert for Cytokine Storm: Immunopathology in COVID-19. Arthritis Rheumatol 2020;72:1059-63. [Crossref] [PubMed]
- Mo Y, Adarkwah O, Zeibeq J, et al. Treatment with Tocilizumab for Patients with Covid-19 Infections: A Case-series Study. J Clin Pharmacol 2021;61:406-11. [Crossref] [PubMed]
- Runfeng L, Yunlong H, Nanshan Z, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res 2020;156:104761 [Crossref] [PubMed]
- Akalın E, Ekici M, et al. Traditional Chinese medicine practices used in COVID-19 (Sars-cov 2/Coronavirus-19) treatment in clinic and their effects on the cardiovascular system. Turk Kardiyol Dern Ars 2020;48:410-24. [PubMed]
- Ren JL, Zhang AH, Wang XJ. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res 2020;155:104743 [Crossref] [PubMed]
- Huang J, Tao G, Liu J, et al. Current Prevention of COVID-19: Natural Products and Herbal Medicine. Front Pharmacol 2020;11:588508 [Crossref] [PubMed]
- Wu XV, Dong Y, et al. Traditional Chinese Medicine as a complementary therapy in combat with COVID-19 - a review of evidence-based research and clinical practice. J Adv Nurs 2021;77:1635-44. [Crossref] [PubMed]
- National Health Commission of the People's Republic of China, Notice of Diagnosis and Treatment Plan for a New Coronary Virus Pneumonia (Trial Version 7). Available online: http://www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm
- Li Q, Wang H, Li X, et al. The role played by traditional Chinese medicine in preventing and treating COVID-19 in China. Front Med 2020;14:681-8. [Crossref] [PubMed]
- Zhang Y L, Zhang W Y, Zhao X Z, et al. Treating COVID-19 by traditional Chinese medicine: a charming strategy? Traditional Medicine Research 2020;5:178-81.
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008;4:682-90. [Crossref] [PubMed]
- Boezio B, Audouze K, Ducrot P, et al. Network-based Approaches in Pharmacology. Mol Inform 2017;36:10. [Crossref] [PubMed]
- Zhang R, Zhu X, Bai H, et al. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol 2019;10:123. [Crossref] [PubMed]
- Saikia S, Bordoloi M. Molecular Docking: Challenges, Advances and its Use in Drug Discovery Perspective. Curr Drug Targets 2019;20:501-21. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Li B, Ma C, Zhao X, et al. YaTCM: Yet another Traditional Chinese Medicine Database for Drug Discovery. Comput Struct Biotechnol J 2018;16:600-10. [Crossref] [PubMed]
- Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 2019;47:D1102-9. [Crossref] [PubMed]
- Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13:6964-82. [Crossref] [PubMed]
- Lipinski CA, Lombardo F, Dominy BW, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3-26. [Crossref] [PubMed]
- Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 2004;1:337-41. [Crossref] [PubMed]
- Thomford NE, Senthebane DA, Rowe A, et al. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. Int J Mol Sci 2018;19:1578. [Crossref] [PubMed]
- Yao ZJ, Dong J, Che YJ, et al. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des 2016;30:413-24. [Crossref] [PubMed]
- Wang J, Zhao S, Liu M, et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv, 2020. doi:
10.1101/2020.02.05.20020545 . - UniProt Consortium T. UniProt. the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Athanasios A, Charalampos V, Vasileios T, et al. Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Curr Drug Metab 2017;18:5-10. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Binns D, Dimmer E, Huntley R, et al. QuickGO: a web-based tool for gene ontology searching. Bioinformatics 2009;25:3045-6. [Crossref] [PubMed]
- Ogata H, Goto S, Sato K,et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 1999;27:29-34.
- Kong R, Yang G, Xue R, et al. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020;36:5109-11. [Crossref] [PubMed]
- Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269-71. [Crossref] [PubMed]
- Shi Y, Zhang X, Mu K, et al. D3Targets-2019-nCoV: a webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm Sin B 2020;10:1239-48. [Crossref] [PubMed]
- Zhang Y, Li Y, Wang X, et al. Herbal plants coordinate COVID-19 in multiple dimensions - An insight analysis for clinically applied remedies. Int J Med Sci 2020;17:3125-45. [Crossref] [PubMed]
- Du HZ, Hou XY, Miao YH, et al. Traditional Chinese Medicine: an effective treatment for 2019 novel coronavirus pneumonia (NCP). Chin J Nat Med 2020;18:206-10. [Crossref] [PubMed]
- Ling CQ. Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-CoV-2). J Integr Med 2020;18:87-8. [Crossref] [PubMed]
- Zhou H, Chen D, Zhang Y, et al. Efficacy and safety of Xiyanping for COVID-2019: A protocol for systematic review and meta-analysis. Medicine (Baltimore) 2020;99:e22962 [Crossref] [PubMed]
- Fan AY, Gu S, Alemi SF. Research Group for Evidence-based Chinese Medicine. Chinese herbal medicine for COVID-19: Current evidence with systematic review and meta-analysis. J Integr Med 2020;18:385-94. [Crossref] [PubMed]
- Ang L, Song E, Lee HW, et al. Herbal Medicine for the Treatment of Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis of Randomized Controlled Trials. J Clin Med 2020;9:1583. [Crossref] [PubMed]
- Xiong X, Wang P, Su K, et al. Chinese herbal medicine for coronavirus disease 2019: A systematic review and meta-analysis. Pharmacol Res 2020;160:105056 [Crossref] [PubMed]
- Luo X, Ni X, Lin J, et al. The add-on effect of Chinese herbal medicine on COVID-19: A systematic review and meta-analysis. Phytomedicine 2021;85:153282 [Crossref] [PubMed]
- Dandara C, Dzobo K, Chirikure S. COVID-19 Pandemic and Africa: From the Situation in Zimbabwe to a Case for Precision Herbal Medicine. OMICS 2021;25:209-12. [Crossref] [PubMed]
- Ma SC, Du J, But PP, et al. Antiviral Chinese medicinal herbs against respiratory syncytial virus. J Ethnopharmacol 2002;79:205-11. [Crossref] [PubMed]
- Gao Y, Liu F, Fang L, et al. Genkwanin inhibits pro-inflammatory mediators mainly through the regulation of miR-101/MKP-1/MAPK pathway in LPS-activated macrophages. PLoS One 2014;9:e96741 [Crossref] [PubMed]
- Sun J, Li X, Jiao K, et al. Albiflorin inhibits the formation of THP-1-derived foam cells through the LOX-1/NF-κB pathway. Minerva Med 2019;110:107-14. [Crossref] [PubMed]
- Park SW, Kwon MJ, Yoo JY, et al. Antiviral activity and possible mode of action of ellagic acid identified in Lagerstroemia speciosa leaves toward human rhinoviruses. BMC Complement Altern Med 2014;14:171. [Crossref] [PubMed]
- Cui Q, Du R, Anantpadma M, et al. Identification of Ellagic Acid from Plant Rhodiola rosea L. as an Anti-Ebola Virus Entry Inhibitor. Viruses 2018;10:152. [Crossref] [PubMed]
- Marín M, María Giner R, et al. Intestinal anti-inflammatory activity of ellagic acid in the acute and chronic dextrane sulfate sodium models of mice colitis. J Ethnopharmacol 2013;150:925-34. [Crossref] [PubMed]
- Morosetti G, Criscuolo AA, Santi F, et al. Ellagic acid and Annona muricata in the chemoprevention of HPV-related pre-neoplastic lesions of the cervix. Oncol Lett 2017;13:1880-4. [Crossref] [PubMed]
- Li H, Yoon JH, Won HJ, et al. Isotrifoliol inhibits pro-inflammatory mediators by suppression of TLR/NF-κB and TLR/MAPK signaling in LPS-induced RAW264.7 cells. Int Immunopharmacol 2017;45:110-9. [Crossref] [PubMed]
- Ge H, Tang H, Liang Y, et al. Rhein attenuates inflammation through inhibition of NF-κB and NALP3 inflammasome in vivo and in vitro. Drug Des Devel Ther 2017;11:1663-71. [Crossref] [PubMed]
- Bourgonje AR, Abdulle AE, Timens W, et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J Pathol 2020;251:228-48. [Crossref] [PubMed]
- Korotkova M, Lundberg IE. The skeletal muscle arachidonic acid cascade in health and inflammatory disease. Nat Rev Rheumatol 2014;10:295-303. [Crossref] [PubMed]
- Consigny PM. Vascular smooth muscle contraction and relaxation: pathways and chemical modulation. J Vasc Interv Radiol 1991;2:309-17. [Crossref] [PubMed]
- Bornstein SR, Dalan R, Hopkins D, et al. Endocrine and metabolic link to coronavirus infection. Nat Rev Endocrinol 2020;16:297-8. [Crossref] [PubMed]
- Zheng YY, Ma YT, Zhang JY, et al. COVID-19 and the cardiovascular system. Nat Rev Cardiol 2020;17:259-60. [Crossref] [PubMed]
- Rabelo LA, Todiras M, Nunes-Souza V, et al. Genetic Deletion of ACE2 Induces Vascular Dysfunction in C57BL/6 Mice: Role of Nitric Oxide Imbalance and Oxidative Stress. PLoS One 2016;11:e0150255 [Crossref] [PubMed]
- Choi MJ, Lee EJ, Park JS, et al. Anti-inflammatory mechanism of galangin in lipopolysaccharide-stimulated microglia: Critical role of PPAR-γ signaling pathway. Biochem Pharmacol 2017;144:120-31. [Crossref] [PubMed]
- Ferroni P, Basili S, Davi G. Platelet activation, inflammatory mediators and hypercholesterolemia. Curr Vasc Pharmacol 2003;1:157-69. [Crossref] [PubMed]
- Keane MP, Strieter RM. Chemokine signaling in inflammation. Crit Care Med 2000;28: N13-26.
- Lee SM, Cheung CY, Nicholls JM, et al. Hyperinduction of cyclooxygenase-2-mediated pro-inflammatory cascade: a mechanism for the pathogenesis of avian influenza H5N1 infection. J Infect Dis 2008;198:525-35. [Crossref] [PubMed]
- Imhof BA, Jemelin S, Ballet R, et al. CCN1/CYR61-mediated meticulous patrolling by Ly6Clow monocytes fuels vascular inflammation. Proc Natl Acad Sci U S A 2016;113:E4847-56. [Crossref] [PubMed]
- Schuliga M. NF-kappaB Signaling in Chronic Inflammatory Airway Disease. Biomolecules 2015;5:1266-83. [Crossref] [PubMed]
- Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185-92. [Crossref] [PubMed]
- Zhu Q, He G, Wang J, Wang Y, et al. Protective effects of fenofibrate against acute lung injury induced by intestinal ischemia/reperfusion in mice. Sci Rep 2016;6:22044. [Crossref] [PubMed]
- Wei X, Xiao YT, Wang J, et al. Sex differences in severity and mortality among patients with COVID-19: evidence from pooled literature analysis and insights from integrated bioinformatic analysis. arXiv preprint arXiv: 2003.13547,2020.
- Mao L, Jin H, Wang M, et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol 2020;77:683-90. [Crossref] [PubMed]
- Gao Y, Yan L, Huang Y, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv, 2020. doi:
10.1101/2020.03.16.993386 . - Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci U S A 2015;112:9436-41. [Crossref] [PubMed]
- Jia Z, Yan L, Ren Z, et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 2019;47:6538-50. [Crossref] [PubMed]
- Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, et al. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta 2013;1828:2026-31. [Crossref] [PubMed]


