
Investigation of the potential mechanism of farnesol in protecting the intestinal epithelium barrier from invasion by Candida albicans via untargeted metabolomics
Introduction
Fungal infections can be life-threatening, and have been shown to cause high morbidity and mortality in immunocompromised and intensive care patients (1,2). Candida albicans (C. albicans) is a common microorganism in the human intestine. Under physiological conditions, when the host has an intact intestinal mucosal barrier and a functioning innate immune system, C. albicans acts as a symbiotic member of the gastrointestinal flora. There exists a homeostasis between the host and the yeast (3). However, when homeostasis is disrupted, C. albicans can enter the bloodstream by invading the intestinal epithelium barrier through microfold cells, resulting in invasive candidiasis and candidemia (4). In recent years, C. albicans infection has emerged as a life-threatening disease (5).
The infections caused by C. albicans are associated with biofilm formation, which is controlled by the quorum sensing molecule farnesol (6,7). It has been reported that farnesol is endogenously generated in C. albicans by enzymatic dephosphorylation of farnesyl diphosphate, and plays a critical role in the physiology of C. albicans by inhibiting hyphal and biofilm formation (8-10). Interestingly, our previous study demonstrated that supplementation of exogenous farnesol that is consistent with the structure and function of farnesol secreted by C. albicans can promote intestinal barrier integrity (11). We speculated that farnesol may protect the intestinal epithelium barrier from invasion by C. albicans by inhibiting hyphal and biofilm formation. However, the underlying mechanisms need to be explored.
Metabolomics has emerged as a promising tool in various fields of human health (12,13). Metabolites are the final products of cell metabolism and regulation, and can act as measures of biochemical status to reflect tissue physiology. Thus, a comparison of different metabolomic profiles can aid in understanding disease mechanisms (14). In this study, we aimed to explore the mechanisms of farnesol on Caco-2 cells, a monolayer model of intestinal epithelial cells, via untargeted metabolomics.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/apm-20-2414).
Methods
Candida strains and cell line
The C. albicans reference strain SC5314 was used, which was grown in yeast extract-peptone-dextrose (YPD) medium containing 2% glucose from glycerol stock cultures for 48 h at 30 °C. Human colorectal cancer epithelial cells (Caco-2) were purchased from Stem Cell Bank, Chinese Academy of Sciences, and grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS) and 1 × non-essential amino acid at 37 °C with 5% CO2.
CCK-8 assay
Caco-2 cells were seeded in a 96-well plate with 2×104 cells per well, and were cultured in DMEM containing 15% FBS to a density of 85–95%. C. albicans at logarithmic growth stage were washed twice with PBS and suspended in serum-free DMEM containing 100, 200, or 300 µM farnesol (Sigma Aldrich, USA) and 0.3% methanol. Then, the Caco-2 culture medium was removed, and 100 µL of C. albicans suspension (8.3×106 cells/mL) was inoculated in each well (4 wells per group). After incubation at 37 °C with 5% CO2 for 1, 2, and 3 h, respectively, the culture medium was removed and the nonadherent cells were rinsed with sterile PBS 3 times. Then, 100 µL DMEM was added followed by the addition of 10 µL CCK-8 to each well for 1 h incubation. The absorbance was detected at 450 nm.
C. albicans biofilm cultivation
Logarithmically grown C. albicans were washed, counted, and suspended in RPMI1640 medium with a cell density of 106 cells/mL. A 96-well plate was inoculated with 100 µL suspension in each well, with 6 wells for repetition. The plate was incubated at 37 °C (5% CO2) for 2 h. The culture medium was then removed, and wells were rinsed with sterile PBS 3 times. The experimental group was treated with 100 µL RPMI1640 containing 200 µmol/L farnesol, and the control group was treated with 100 µL RPMI1640. The morphology of C. albicans was observed and the culture medium was changed every day.
Metabolite extraction
There were 4 treatment groups: Caco-2 + C. albicans (group 1), Caco-2 (group 2), Caco-2 + C. albicans + farnesol (group 3), and a quality control (QC group) for metabolite extraction. The sample size was 6 for the 3 experimental groups, and 3 for the QC group. In detail, 40 mg of samples from each group was dissolved in 1 mL of methanol:acetonitrile:H2O (2:2:1, v/v/v), followed by sonication for 30 min at 4 °C. Protein was precipitated at –20 °C for 1 h then centrifuged at 13,000 rpm at 4 °C for 15 min. The supernatants were lyophilized and stored at −80 °C. Dried samples were re-dissolved in 100 µL acetonitrile:H2O (1:1, v/v) and centrifuged at 14,000 rpm for 15 min.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
The samples were placed in a 4 °C autosampler and separated using a HILIC chromatographic column with the Agilent 1290 Infinity LC UHPLC system at 0.3 mL/min with a column temperature of 25 °C. The eluent consisted of A: 25 mM ammonium acetate and 25 mM ammonia, and B: acetonitrile. Chromatographic gradient elution procedures were 95% B (0–0.5 min); from 95% B to 65% B (0.5–7 min); from 65% B to 40% B (7–8 min); 40% B (8–9 min); 40% B to 95% B (9–9.1 min); 95% B (9.1–12 min).
Electrospray ionization (ESI) was used to detect positive and negative ions. The samples were analyzed using a Triple TOF 5,600 mass spectrometer (AB SCIEX) after being separated by ultra performance liquid chromatography (UPLC). ESI conditions were set as follows: Ion Source Gas1 60, Ion Source Gas2 60, Curtain gas 30, source temperature 600 °C, IonSapary Voltage Floating ±5,500 V (positive and negative ion mode), TOF MS scan m/z range 60–1,200 Da, product ion scan m/z range 25–1,200 Da, TOF MS scan accumulation time 0.15 s/spectra, product ion scan accumulation time 0.03 s/spectra, secondary mass spectrometry was acquired by information-dependent acquisition (IDA) with a high-sensitivity model, declustering potential ± 60 V, collision energy 30 eV, IDA excluded isotopes within 4 Da, and candidate ions monitored per cycle was 6.
Data processing
The original data were converted into .mzXML format by ProteoWizard, then the peak alignment, retention time correction, and peak area extraction were performed using XCMS. Accurate mass number matching (<25 ppm) and secondary spectrum matching methods were used for metabolite structure identification. The metabolites in the positive and negative modes were mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) IDs using MetaboAnalyst (15).
Data analysis
In the positive or negative ion mode, metabolites that could be matched to KEGG IDs were extracted and normalized using the Pareto-scaling method in METAGENassist (16). Principal component analysis (PCA) was carried out to observe the overall sample distribution and the stability of the experimental batch using the prcomp function in R. Partial least squares discriminant analysis (PLS-DA) was applied to identify the overall differences in the metabolic profiles among groups, and the differential metabolites associated with disease/grouping were determined using the mixOmics package in R.
Differential metabolite screening
The differential metabolites of group 1 vs. group 1 and group 3 vs. group 1 in the positive and negative ion mode were identified. During the identification of differential metabolites, the variable importance parameter (VIP) value of multivariate statistical analysis was calculated using the ropls package of R. The metabolites with P value <0.05, |log2 fold change (FC)| >0.585 and VIP >1 were considered as differential metabolites.
KEGG enrichment analysis
To elucidate the biological significance of differences in metabolites, compounds, and gene expression, the Integrated Molecular Pathway-Level Analysis (IMPaLA) tool was used to select significant pathways enriched by differential metabolites, with the KEGG database as the background. Significance thresholds were set as count ≥2 and P<0.05.
Results
Effects of different concentrations of farnesol on the co-culture of C. albicans and Caco-2 cells
With the increase in the concentration of farnesol, the cytotoxicity of farnesol also increased. Some of the Caco-2 cells were exfoliated after 200 µM farnesol treatment, and most Caco-2 cells were exfoliated when treated with 300 µM farnesol. There was no significant effect of 100 µM farnesol on Caco-2 cells (Figure 1A). Additionally, after 2 or 3 h of incubation, farnesol significantly reduced the adhesion of C. albicans (Figure 1B).
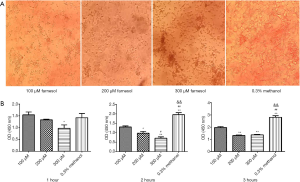
Effect of farnesol on the formation of C. albicans biofilm
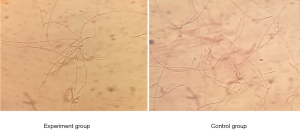
After being cultured in RPMI for 2 h at 37 °C with 5% CO2, the cells began to grow short hyphae. The cells in experimental group were then cultured with RPMI containing 200 µM farnesol. After 48 h of further culture, only monolayer biofilm was formed in the experimental group, and mycelia could be clearly observed. In the control group, multilayer thick biofilm was found, and single hypha were difficult to observe (Figure 2). Thus, treatment with 200 µmol/L farnesol for 48 h significantly inhibited the formation of C. albicans biofilm, with developed pseudohypha.
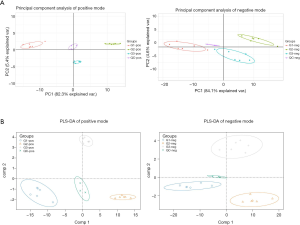
Data analysis PCA and PLS-DA analysis of the metabolic profiles
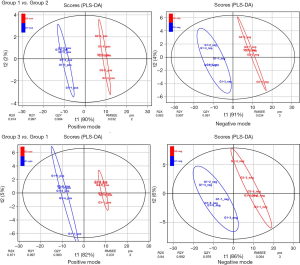
There were 190 metabolites with annotation information in the positive ion peak, and 109 were mapped to KEGG IDs. In the negative ion peak, there were 180 metabolites with annotation information, 123 of which were mapped to KEGG IDs. PCA and PLS-DA analysis results showed that QC samples had only a small deviation in the positive and negative ion mode, indicating the stability and reliability of the experimental process. The other 3 groups of data showed an obvious trend of separation, indicating that the metabolic profiles of the 3 groups were different, and the data were worthy of further analysis (Figure 3A,B). Additionally, we also performed PLS-DA analysis for group 1 vs. group 2 and group 3 vs. group 1, respectively, and obtained the VIP value as one of the indicators for selecting differential metabolites. As shown in Figure 4, in the positive and negative mode, different groups presented a clear separation trend, and both R2Y (the interpretation rate of the model to the grouping) and Q2Y (predictive ability of the model) were close to 1, indicating that the interpretation rate and predictive ability of the model for grouping were better, and the differential analysis results were credible.
Differential metabolite identification
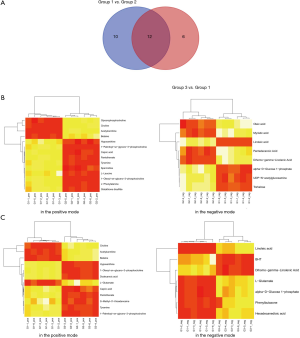
The union of the differential metabolites in the positive and negative modes was taken as the differential metabolites. Finally, based on P value <0.05, |log2FC| >0.585 and VIP >1, a total of 22 differential metabolites were identified in group 1 vs. group 2, including 1-oleoyl-sn-glycero-3-phosphocholine (downregulated), 1-palmitoyl-sn-glycero-3-phosphocholine (downregulated), acetylcarnitine (downregulated), alpha-D-glucose 1-phosphate (upregulated), betaine (downregulated), capric acid (downregulated), choline (downregulated), dihomo-gamma-linolenic acid (downregulated), glutathione disulfide (downregulated), glycerophosphocholine (downregulated), hypoxanthine (upregulated), L-leucine (downregulated), L-phenylalanine (downregulated), linoleic acid (downregulated), myristic acid (upregulated), oleic acid (upregulated), pantothenate (downregulated), pentadecanoic acid (upregulated), spermidine (downregulated), trehalose (upregulated), tyramine (downregulated), and UDP-N-acetylglucosamine (upregulated). In addition, there were 18 differential metabolites identified in group 3 vs. group 1, including 1-oleoyl-sn-glycero-3-phosphocholine (upregulated), 1-palmitoyl-sn-glycero-3-phosphocholine (upregulated), acetylcarnitine (upregulated), alpha-D-glucose 1-phosphate (downregulated), betaine (upregulated), BHT (downregulated), capric acid (upregulated), choline (upregulated), dihomo-gamma-linolenic acid (upregulated), dodecanoic acid (upregulated), hexadecanedioic acid (downregulated), hypoxanthine (downregulated), L-glutamate (downregulated), linoleic acid (upregulated), pantothenate (upregulated), phenylbutazone (downregulated), S-methyl-5'-thioadenosine (upregulated), and tyramine (upregulated). Venn analysis showed that there were 12 common differential metabolites between the 2 comparison groups, including linoleic acid, capric acid, choline, dihomo-gamma-linolenic acid, alpha-D-glucose 1-phosphate, 1-palmitoyl-sn-glycero-3-phosphocholine, acetylcarnitine, tyramine, hypoxanthine, betaine, 1-oleoyl-sn-glycero-3-phosphocholine, and pantothenate (Figure 5A). Moreover, the obtained differential metabolites were subjected to bidirectional hierarchical clustering based on the metabolic data extracted from the normalized data. As shown in Figure 5B,C, the differential metabolites were divided into 2 distinct groups by clustering.
Pathway enrichment analysis
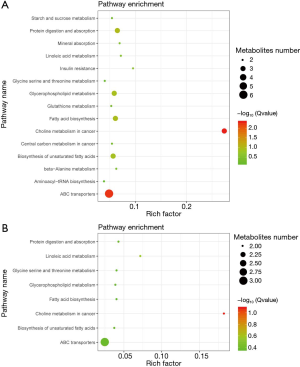
Using the IMPaLA tool, 15 pathways were identified based on the differential metabolites in group 1 vs. group 2, including ABC transporters, choline metabolism in cancer, protein digestion and absorption, fatty acid biosynthesis, glycerophospholipid metabolism, biosynthesis of unsaturated fatty acids, insulin resistance, linoleic acid metabolism, mineral absorption, starch and sucrose metabolism, beta-alanine metabolism, central carbon metabolism in cancer, glutathione metabolism, aminoacyl-tRNA biosynthesis, and glycine serine and threonine metabolism (Figure 6A). Furthermore, 8 pathways were enriched by the differential metabolites in group 3 vs. group 1, including ABC transporters, choline metabolism in cancer, linoleic acid metabolism, protein digestion and absorption, fatty acid biosynthesis, glycine serine and threonine metabolism, glycerophospholipid metabolism, and biosynthesis of unsaturated fatty acids (Figure 6B).
Discussion
C. albicans is a polymorphic fungal pathogen which colonizes the human gastrointestinal mucosal tissues, and has effects on the intestinal mucosal barrier (17). C. albicans can serve as a pathogen in various infections and cause mucosal diseases (18). Farnesol, a quorum sensing molecule, can prevent the hyphal formation of C. albicans, and it is therefore considered to play a key role in the pathogenic processes of C. albicans (19,20). In this study, we also found that farnesol inhibited the hyphal formation and biofilm formation of C. albicans. Through metabolomics analysis, we found that C. albicans altered the metabolism of the intestinal epithelial cell model, while farnesol reversed these metabolic changes.
In the present study, by comparing the differential metabolites between group 1 (Caco-2 + C. albicans) and group 2 (Caco-2), 22 significant differential metabolites were identified, such as acetylcarnitine (downregulated in group 1). It has been reported that acetylcarnitine, a zwitterionic surfactant, has a unique structure which can perturb the rat jejunum and decrease membrane resistance in the rat colon and in Caco-2 cell monolayers (21,22). Additionally, acetylcarnitine is able to open intestinal tight junctions by affecting certain claudin subtypes (23). Interestingly, acetylcarnitine was upregulated after farnesol treatment, which suggests that farnesol may protect the intestinal epithelium barrier from the invasion of C. albicans by regulating acetylcarnitine.
Intestinal functions are usually affected by intestinal morphology such as villus area, villus height, and crypt depth (24). Maternal dietary linoleic acid supplementation has been shown to increase villus area and villus height compared with controls (25). A previous study indicated that linoleic acid deficiency in rats resulted in significantly lower villus height in the ileum compared to controls (26). The above findings suggest the important role of linoleic acid in intestinal health. In this study, linoleic acid was downregulated in the Caco-2 + C. albicans group compared with the Caco-2 group, which showed involvement of the linoleic acid metabolism pathway. This indicates that C. albicans may impair intestinal health via decreasing linoleic acid metabolism. Nevertheless, in the Caco-2 + C. albicans + farnesol group, linoleic acid was upregulated compared with the Caco-2 + C. albicans group, which suggested that farnesol could improve the intestinal epithelium barrier by increasing linoleic acid metabolism.
Glutathione, a free radical-scavenging compound, is a major antioxidant in intestinal epithelial cells and plays an important role in intestinal barrier function (27-29). Relatively high concentrations of glutathione have been detected in the intestinal epithelium (30). A previous study reported that Salmonella infection increases the sensitivity of epithelial cells to oxidative damage by reducing glutathione levels in mouse ileal cells (31). In this study, spermidine and glutathione disulfide, 2 downregulated metabolites in the Caco-2 + C. albicans group compared with the Caco-2 group, were enriched in glutathione metabolism. We speculated that C. albicans may also cause oxidative damage in intestinal epithelial cells by decreasing glutathione metabolism.
L-glutamate is a major oxidative fuel for the gastrointestinal tract and is also the preferred energy source for the gut (32,33). Studies in mammals have indicated that approximately 96% of enteral L-glutamate is metabolized in the small intestine during the first pass (34,35). L-glutamate is essential for maintaining antioxidative responses and intestinal mucosa integrity (36). Jiao et al. (37) reported that L-glutamate could maintain intestinal barrier function in diquat-challenged enterocytes by increasing the expression level of tight junction proteins. In this study, L-glutamate was downregulated in the Caco-2 + C. albicans + farnesol group compared with the Caco-2 + C. albicans group. We speculated that farnesol may promote the metabolism of L-glutamate to maintain intestinal barrier function in the C. albicans-affected intestinal barrier.
In conclusion, our study suggests that C. albicans may damage the intestinal barrier by affecting the metabolism of acetylcarnitine, linoleic acid, and glutathione. Farnesol may protect the intestinal epithelium barrier from invasion of C. albicans by regulating the metabolism of acetylcarnitine, linoleic acid, and L-glutamate.
Acknowledgments
Funding: Health and Family Planning Commission Research Project Youth Project of Shanghai [20174Y0061], Natural Science Foundation of Minhang District Science and Technology Commission of Shanghai [2019MHZ098].
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/apm-20-2414
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-2414
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-2414). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pongrácz J, Juhász E, Iván M, et al. Significance of yeasts in bloodstream infection: Epidemiology and predisposing factors of Candidaemia in adult patients at a university hospital (2010-2014). Acta Microbiol Immunol Hung 2015;62:317-29. [Crossref] [PubMed]
- Li Y, Du M, Chen LA, et al. Nosocomial Bloodstream Infection Due to Candida spp. in China: Species Distribution, Clinical Features, and Outcomes. Mycopathologia 2016;181:485-95. [Crossref] [PubMed]
- Schulze J, Sonnenborn U. Yeasts in the gut: from commensals to infectious agents. Dtsch Arztebl Int 2009;106:837-42. [PubMed]
- Tong Y, Tang J. Candida albicans infection and intestinal immunity. Microbiol Res 2017;198:27-35. [Crossref] [PubMed]
- Yan L, Yang C, Tang J. Disruption of the intestinal mucosal barrier in Candida albicans infections. Microbiol Res 2013;168:389-95. [Crossref] [PubMed]
- Polke M, Leonhardt I, Kurzai O, et al. Farnesol signalling in Candida albicans–more than just communication. Crit Rev Microbiol 2018;44:230-43. [Crossref] [PubMed]
- Kong EF, Tsui C, Kucharíková S, et al. Modulation of Staphylococcus aureus Response to Antimicrobials by the Candida albicans Quorum Sensing Molecule Farnesol. Antimicrob Agents Chemother 2017;61:e01573-17. [Crossref] [PubMed]
- Hornby JM, Jensen EC, Lisec AD, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 2001;67:2982-92. [Crossref] [PubMed]
- Sato T, Watanabe T, Mikami T, et al. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol Pharm Bull 2004;27:751-2. [Crossref] [PubMed]
- Uppuluri P, Mekala S, Chaffin WL. Farnesol-mediated inhibition of Candida albicans yeast growth and rescue by a diacylglycerol analogue. Yeast 2007;24:681-93. [Crossref] [PubMed]
- Fang Y, Wu C, Wang Q, et al. Farnesol contributes to intestinal epithelial barrier function by enhancing tight junctions via the JAK/STAT3 signaling pathway in differentiated Caco-2 cells. J Bioenerg Biomembr 2019;51:403-12. [Crossref] [PubMed]
- Cortés M, Pareja E, Castell JV, et al. Exploring mass spectrometry suitability to examine human liver graft metabonomic profiles. Transplant Proc 2010;42:2953-8. [Crossref] [PubMed]
- Zhu H, Wang Z, Wu Y, et al. Untargeted metabonomics reveals intervention effects of chicory polysaccharide in a rat model of non-alcoholic fatty liver disease. Int J Biol Macromol 2019;128:363-75. [Crossref] [PubMed]
- García-Cañaveras JC, Donato MT, Castell JV, et al. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J Proteome Res 2011;10:4825-34. [Crossref] [PubMed]
- Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486-W494. [Crossref] [PubMed]
- Arndt D, Xia J, Liu Y, et al. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 2012;40:W88-95. [Crossref] [PubMed]
- Mao X, Qiu X, Jiao C, et al. Candida albicans SC5314 inhibits NLRP3/NLRP6 inflammasome expression and dampens human intestinal barrier activity in Caco-2 cell monolayer model. Cytokine 2020;126:154882. [Crossref] [PubMed]
- Egbe NE, Dornelles TO, Paget CM, et al. Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae. Microbial Cell 2017;4:294. [Crossref] [PubMed]
- Wongsuk T, Pumeesat P, Luplertlop N. Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol 2016;56:440-7. [Crossref] [PubMed]
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence 2013;4:119-28. [Crossref] [PubMed]
- Tomita M, Doi N, Hayashi M. Effects of acylcarnitines on efflux transporting system in Caco-2 cell monolayers. Eur J Drug Metab Pharmacokinet 2010;35:1-7. [Crossref] [PubMed]
- Tomita M, Doi N, Hayashi M. Effects of lauroylcarnitine and palmitoylcarnitine on transport of ranitidine in human intestinal epithelial Caco-2 cell monolayers. Organ Biology 2008;15:377-85.
- Doi N, Tomita M, Hayashi M. Absorption Enhancement of Acylcarnitine through Changes in Tight Junction Protein in Caco-2 Cell Monolayers. Drug Metab Pharmacokinet 2011;26:162-70. [Crossref] [PubMed]
- Li J, Yin L, Wang L, et al. Effects of vitamin B6 on growth, diarrhea rate, intestinal morphology, function, and inflammatory factors expression in a high-protein diet fed to weaned piglets1. J Anim Sci 2019;97:4865-74. [Crossref] [PubMed]
- Xu Q, Wen J, Wang X, et al. Maternal dietary linoleic acid altered intestinal barrier function in domestic pigeons (Columba livia). Br J Nutr 2020. Epub ahead of print. [Crossref] [PubMed]
- Thomson AB, Keelan M, Clandinin MT, et al. Dietary fat selectively alters transport properties of rat jejunum. J Clin Invest 1986;77:279-88. [Crossref] [PubMed]
- van Ampting MT, Schonewille AJ, Vink C, et al. Intestinal barrier function in response to abundant or depleted mucosal glutathione in Salmonella-infected rats. BMC Physiol 2009;9:6. [Crossref] [PubMed]
- Meister A. Glutathione metabolism and its selective modification. J Biol Chem 1988;263:17205-8. [Crossref] [PubMed]
- Zhou X, Zhang Y, He L, et al. Serine prevents LPS-induced intestinal inflammation and barrier damage via p53-dependent glutathione synthesis and AMPK activation. Journal of Functional Foods 2017;39:225-32. [Crossref]
- Kelly FJ. Glutathione content of the small intestine: regulation and function. Br J Nutr 1993;69:589-96. [Crossref] [PubMed]
- Mehta A, Singh S, Ganguly NK. Impairment of intestinal mucosal antioxidant defense system during Salmonella typhimurium infection. Dig Dis Sci 1998;43:646-51. [Crossref] [PubMed]
- Hou Y, Wu G. Nutritionally nonessential amino acids: a misnomer in nutritional sciences. Adv Nutr 2017;8:137-9. [Crossref] [PubMed]
- Brosnan JT, Brosnan ME. Glutamate: a truly functional amino acid. Amino Acids 2013;45:413-8. [Crossref] [PubMed]
- Burrin DG, Stoll B. Metabolic fate and function of dietary glutamate in the gut. Am J Clin Nutr 2009;90:850S-6S. [Crossref] [PubMed]
- Wu G. Intestinal mucosal amino acid catabolism. J Nutr 1998;128:1249-52. [Crossref] [PubMed]
- König J, Wells J, Cani PD, et al. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol 2016;7:e196. [Crossref] [PubMed]
- Jiao N, Wu Z, Ji Y, et al. L-glutamate enhances barrier and antioxidative functions in intestinal porcine epithelial cells. J Nutr 2015;145:2258-64. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


