Utilizing network pharmacology to explore the underlying mechanism of Qiangzhi decoction in treating Tourette’s syndrome
Introduction
Tourette’s syndrome (TS), also known as Gilles de la Tourette’s syndrome, is a neurodevelopmental condition characterized by tics and is usually accompanied by behavioral symptoms (1). Individuals with TS have multiple vocal and motor tics that may trace back to their childhood, and the tics are ongoing. According to observed signs and symptoms of TS in the clinic, including common motor tics including facial grimacing, head/neck movements, eye blinking, shoulder/upper body movements, and common vocal tics such as coughs, sniffs, grunts, and throat-clearing noises. These signs and symptoms can be restrained for periods; however, intense mood swings, including excitement, anxiety, or stress, often worsen tics (2). Severe TS with noticeable tics significantly affects patients’ daily and social life, leading to mental disorders, including inferiority complex, anxiety, depression, and rebellion in specific individuals (3). At present, pharmacological treatment options available to treat TD are using oral synthetic drugs; Clonidine (4), Guanfacine (5), Haloperidol (6), Risperidone (7), and Aripiprazole (8). Non-pharmacological treatments, including habit reversal therapy (9), are in practice. However, most pharmacological treatments available for TS alleviate only the symptoms but not intrinsic causes. These treatments also cause unwanted side effects, including drug dependence. Thus, safer and more effective therapeutic options are needed to treat TS.
Traditional Chinese herbal medicine (TCHM) has been adopted widely for centuries to treat several diseases. Many herbal medicines derived from natural sources have shown to display only minimal side effects (10). Many TCHMs have already been used in the treatment of TS, and meta-analysis supports the efficacy and safety of TCHM alone and TCHM + Western medicine (WM) in TS patients (11). Therefore, it would be helpful to identify the most effective TCHM compounds to treat TS. The traditional Chinese herbal medicine compound prescription (TCHMCP or Chinese medicine prescription) is the main form of Chinese medicine. It is based on the dialectical thought point of view, to choose the appropriate quantitative compatibility of herbal medicines, which is the concrete embodiment of the theory of dialectical treatment of traditional Chinese medicine. It is of great significance to promote TCHMCP to the international community. In order to give full play to the advantages and benefits of TCHMCP, it is necessary to study Chinese herbal medicine compound prescription by modern methodology and technology, clarify the material basis (effective components) and action principle of compound prescription, which is the ultimate goal of modernization research on Chinese herbal compound prescriptions, and this could also provide theoretical support for the further research and development of new drugs.
Earlier studies and clinical observations have revealed the feasibility and effectiveness of Qiangzhi decoction (QD) in treating TS, significantly reducing tics’ frequency during the disease. According to the Zhiyi Dialectical Theory (12) and Traditional Chinese medicine (TCM) theory, QD is classified as a curative TCHM formula and is being used to treat several diseases for decades. The QD contains six Chinese herbal medicines, namely renshen (RS) derived from Panax Ginseng, C. A. Mey, bajitian (BJT) derived from Morindae Officinalis Radix, muxiang (MX) derived from Aucklandiae Radix, shanyao (SY) derived from Rhizoma Dioscoreae, fuling (FL) derived from Poria Cocos (Schw.) Wolf and baiziren (BZR) derived from Platycladi Semen. This study aims to predict the potential pharmacological mechanisms of QD action and its ingredients in treating TS using the network pharmacology method.
Network pharmacology is a powerful and helpful method that integrates bioinformatics, network biology, chemo-informatics, traditional pharmacology, and network analysis (13). The network pharmacology method is being established as an alternative method for elucidating the potential mechanisms and active compounds of TCHM, and its principles are in agreement with those in TCM theory. In this study, we have identified the molecular targets for the compounds in QD and associated them with those targets in TS’s etiology.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/apm-20-2158).
Methods
Construction of a database of the components of QD
Figure 1 shows a schematic representation of the method adopted in this study, using network pharmacology to predict QD molecular mechanisms in treating TS. The six herbs in QD are RS, BJT, MX, SY, FL, and BZR. The data regarding the compounds present in RS, BJT, MX, SY, FL and BZR were derived from TCMSP (the Traditional Chinese Medicine Systems Pharmacology database) (14), and TCM (the Traditional Chinese Medicine Databases @ Taiwan) (15). Five hundred fifty-eight compounds were identified in QD, including 190 compounds from RS, 174 compounds from BJT, 106 compounds from MX, 34 compounds from FL, 71 compounds from SY, and 28 compounds BZR.
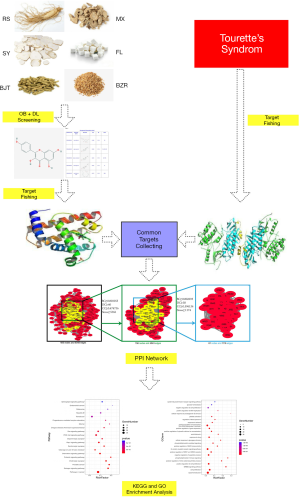
Screening of the active ingredients in QD
The 558 potential compounds from QD were filtered using two models of Absorption, Distribution, Metabolism, and Excretion (ADME) in which drug-likeness (DL) and oral bioavailability (OB) were integrated. Drug-likeliness is a qualitative value applied in drug design to decide whether the compound is drug-like and is used as a parameter to optimize and improve its pharmaceutical and pharmacokinetic properties (16,17). OB represents the relative amount of orally administered drug that reaches the blood circulation. The compounds with values of DL ≥0.18 and OB ≥30%, from the information available in the TCMSP database and the literature, were selected for further analysis (18). The putative targets of the compounds in QD were identified from the STITCH (19) and PubChem databases (http://pubchem.ncbi.nih.gov/) (20). The targets with inadequate information were excluded.
Target genes related to the compounds
Swiss Target Prediction (21) and TCMSP databases were used to identify the locks on identifying the relevant target genes associated with QD compounds. According to former research (22), the target genes with a confidence score value of >0.6 in Swiss Target Prediction were selected for further studies. The TCMSP is a curated database of small molecules’ chemical structure, biological activities, and other relevant information. The data in TCMSP is usually experimentally verified. We entered all the active biomolecules identified in the present study into the Swiss Target Prediction and TCMSP databases by drawing the chemical structures and selecting Homo -sapiens as the species set. The gene information, including the gene ID, organism, and name, was confirmed by the Uniprot protein sequence resource (23).
Target genes related to TS
Information on TS-associated target genes was retrieved from the following resources. GeneCards, a comprehensive database, has all the information regarding predicted and annotated genes (24). It has identified 694 genes associated with TS. DrugBank (25) is a comprehensive online database that supplies pharmacological and biochemical information on drugs, molecular targets, and action mechanisms. Forty genes associated with TS were identified. Online Mendelian Inheritance in Man® (OMIM) database) (26) is a comprehensive resource of human gene targets and genetic phenotypes, from which 200 genes related to TS were selected. DisGeNET (27) is a database for exploring certain human diseases and their genes, from which TS related 96 genes were selected. After removing the duplicates, 778 targets linked to TS were selected for further study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Construction of pharmacological network method of pharmacological network construction
The pharmacological network was constructed, using the following processes: (I) a compound-target network was established by linking compounds and predicted targets with a degree of >3.0; (II) A QD target-TS target interaction network [protein-protein interaction (PPI) was established]; and (III) Target-pathway network was established. The pathway information about the selected targets was retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The pharmacological network was visualized using Cytoscape3.6.0, an open-source program for visualizing network analysis (28).
The topological feature set definition of pharmacological network
We selected four parameters to estimate each node’s topological features in the QD target-TS target interaction network: (I). ‘Degree’ (29); (II) ‘Betweenness Centrality’ (30); (III) ‘Closeness Centrality’ (31); (IV) ‘The Stress’. The level of the four parameters is the topological importance of the nodes in the pharmacological network. A node was defined as a major node if all the four topological features were higher than the corresponding median levels (32).
Pathway enrichment analysis
Database for Annotation, Visualization, and Integrated Discovery (DAVID) (33) was used in this study for gene ontology (GO) enrichment analysis. To conduct the pathway enrichment analysis, we applied the KEGG (34) data retrieved from DAVID.
Statistical analysis
The pathway information about the selected targets was retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The topological data were analysed by Cytoscape version 3.6.0. for Windows, WPS Office version 11.1.0.9739-Release for Windows. Pathway enrichment analysis was run by Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8. All the relevant data analysis results are presented in “Results” Section below.
Results
Screening for the active compounds of QD
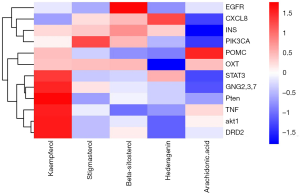
From the six active components of QD; RS, BJT, MX, FL, SY, and BZR; 558 compounds were retrieved from TCMSP and TCM databases. Out of 558 compounds, 190 (31.5%) compounds from RS, 174 (28.9%) compounds were from BJT, 106 (17.6%) compounds were from MX, 34 (5.6%) compounds were from FL, 71 (11.8%) compounds were from SY, and 28 (4.6%) compounds were from BZR. Seventy-eight compounds meet the OB (≥30%) and DL (≥0.18). The network of the Herbs-Compounds was constructed, and the top five compounds were found with edge betweenness. The five compounds are kaempferol (MOL000422, OB =41.88, DL =0.24, found in RS), stigmasterol (MOL000449, OB =43.83, DL =0.76, found in RS, SY, MX), beta-sitosterol (MOL000359, OB =36.91, DL =0.75, found in BJT, RS), hederagenin (MOL000296, OB =36.91, DL =0.75, found in FL), and arachidonic acid (MOL001439, OB =45.57, DL =0.20, found in BZR) (Table 1).
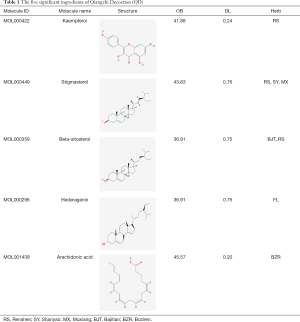
Full table
Target analysis and prediction
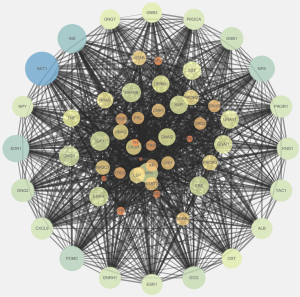
The STITCH, Swiss Target Prediction, and PubChem databases were used in this process for target fishing on the 558 active compounds from chemical similarity, gaining 588 relevant targets, among which there were 313 in BJT, 82 in MX, 454 in RS, 71 in FL, 221 in SY, and 71 in BZR. Seven hundred seventy-eight targets were identified for TS from the database; GeneCards, DrugBank, OMIM database, DisGeNET. Fifty-four of these targets were the same as those predicted molecular targets of the QD compounds and considered the potential targets for QD in TS (Figures 2,3).
The PPI network of the above 54 targets was constructed using the STRING database. Four hundred six human proteins associated with the 54 targets were identified and resulted in 406 nodes and 9,348 edges. The PPI network topology was analyzed on the four parameters: “betweenness”, “degree”, “closeness”, and “stress”. The targets with the value greater than the median values of each parameter were targets through which QD could show a therapeutic effect in TS. In the first screening, the threshold values were set at closeness ≥0.474736, degree ≥40, stress ≥5,366, and betweenness ≥0.001055. It resulted in 156 hub nodes and 4,043 edges. The 156 hub nodes were further by setting the threshold values: closeness ≥0.596154, degree ≥50, stress ≥1,174, and betweenness ≥0.002695. It results in 61 big hub nodes and 1,116 edges (Figure 4). The entire process of the topological feature analysis of the PPI network is shown in Figure 5. For the degree value in descending order and previous research studies on TS, we eventually selected 14 core targets: Akt1 (degree =116), INS (degree =114), CXCL8 (degree =86), POMC (degree =84), GNG2 (degree =82), EGFR (degree =81), PIK3CA (degree =78), GNG7 (degree =76), GNG3 (degree =75), TNF (degree =74), OXT (degree =67), STAT3 (degree =67), DRD2 (degree =67), Pten (degree =62) (Table 2).

Full table
The main active ingredient—target molecular docking
It is generally considered that the lower the conformational stability of ligand receptor binding, the greater the possibility of interaction. Based on former study, a docking score greater than 4.25 indicates a certain binding activity between the docking molecule and the target, values greater than 5.0 indicate good binding activity between the docking molecule and the target (35).
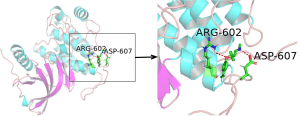
The five compounds, kaempferol (MOL000422), stigmasterol (MOL000449), beta-sitosterol (MOL000359), hederagenin (MOL000296), and arachidonic acid (MOL001439) are the most important active ingredients which have the highest degree in the network of the Herbs-Compounds of QD. The Ledock (Ledock is a cross platform (win, Linux, Mac OS) molecular docking software developed by Dr. Zhao Hongtao of the University of Zurich. It shows strong advantages in analysis speed and accuracy.) was used to verify the molecular docking of the five main medicinal ingredients and the 14 core targets. The docking results showed that the average docking score of the chemical components, i.e., kaempferol, stigmasterol, beta-sitosterol, hederagenin, and arachidonic acid with the targets, i.e., Akt1, INS, CXCL8, POMC, GNG2, EGFR, PIK3CA, GNG7, GNG3, TNF, OXT, STAT3, DRD2, and Pten, was 4.44. Arachidonic acid and POMC have a max docking score of 6.35 (Figure 6). This docking analysis showed that kaempferol, stigmasterol, beta-sitosterol, hederagenin, and arachidonic acid have good binding activity with Akt1, POMC, PIK3CA, GNG2, GNG3, GNG7, OXT, STAT3 and Pten (Figure 7).
KEGG pathway enrichment and GO biological analysis process
The 61 targets retrieved from the STRING database were given as input in DAVID v6.8 for enrichment analysis, with screening threshold set at FDR <0.01. The analysis resulted in 48 GO items and 79 related KEGG pathways used for further analysis.
KEGG pathway enrichment analysis
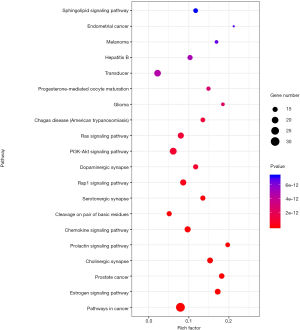
For further revelations of the potential mechanism of QD ion TS’s pharmacological effect, the KEGG pathway enrichment analysis was performed on the 61 targets. Seventy-nine pathways were screened with the threshold value set at FDR <0.01. Figure 8 shows the top 20 pathways of KEGG enrichment, and they are pathways in cancer, Estrogen signaling pathway, Prostate cancer, Cholinergic synapse, Prolactin signaling pathway, Chemokine signaling pathway, Cleavage on pair of basic residues, Serotonergic synapse, Rap1 signaling pathway, Dopaminergic synapse, PI3K-Akt signaling pathway, Ras signaling pathway,
GO biological process enrichment analysis
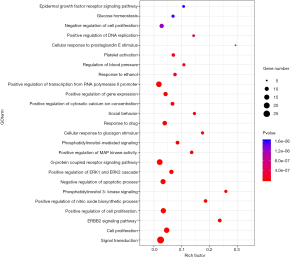
The top 48 biological processes were selected on FDR values after sorting the 466 items, which involved cell proliferation, signal transduction, and gene expression. The fundamental processes involved in cell proliferation are negative regulation of cell proliferation (GO number: 0008285), positive regulation of cell proliferation (GO number: 0008284), positive regulation of smooth muscle cell proliferation (GO number: 0048661). The main processes involved in signal transduction are signal transduction (GO number: 0007165), Ras protein signal transduction (GO number: 0007265), cell-cell signaling (GO number: 0007267), protein kinase B signaling (GO number: 0043491), phosphatidylinositol 3-kinase signaling (GO number: 0014065), G-protein-coupled receptor signaling pathway (GO:0007186). The fundamental processes involved in gene expression are positive regulation of gene expression (GO number: 0010628), positive regulation of DNA replication (GO number: 0045740), positive regulation of transcription from RNA polymerase II promoter (GO number: 0045944). The analysis suggested that QD could show a therapeutic effect in TD via cell proliferation, signal transduction, and gene expression, and these three processes produce a synergistic effect. Figure 9 shows the top 20 biological processes of GO enrichment.
Discussion
TS is a neuro-developmental condition with multiple motor tics and vocal tics, which begin in childhood and are usually accompanied by behavioral symptoms. We noticed that the QD could alleviate the symptoms in TS patients. We postulate that QD activity in TS arises from the compounds present in it and mediated through multiple targets, pathways, and modes. In this study, a network pharmacology approach was adopted to predict QD’s potential mechanisms in TS.
With the Herbs-Compounds network, five hub compounds, kaempferol, stigmasterol, beta-sitosterol, hederagenin, and arachidonic acid, were identified as potent compounds responsible for QD’s therapeutic effect. Kaempferol is a bioflavonoid found in many vegetables and fruits (36). Kaempferol has shown many pharmacological activities; neuroprotective, antioxidant, anti-inflammatory, anti-apoptotic, and anti-estrogenic (37). Hederagenin is a triterpeneis and is present in many herbal medicines, including BZR, and is reported to have anti-inflammatory, anti-tumor, and antidepressant (38) activities.
The results from the Target-Pathway network analysis and GO analysis, the potential molecular mechanisms of QD in TS treatment, are associated with the dopamine system, inflammation, infection, and miRNA pathway.
Dopamine system
The dopamine dysfunction is associated with tics symptoms in TS (39). The research studies have proven that hypersensitization of dopamine receptors, dysfunction of presynaptic dopamine neurons, dopaminergic hyperinnervation, and over-activation of dopamine transporters occur in TS. These events are reported to cause low tonic but high phasic dopamine. TS treatment options are acting through either reducing phasic dopamine, or augmenting tonic dopamine, or both (39). One earlier study reported that dopamine regulations could affect insulin signaling, and these effects could be observed in the pancreas and central nervous system. Insulin also influences dopamine signaling reciprocally with specific dopamine reuptake effects from the synapse (40) and dopaminergic signaling (41). The PTEN/Akt signaling pathway takes part in many neurological diseases. Inhibition of striatal PTEN in vivo leads to motor dysfunction and impaired dopamine responses, particularly D2 receptor agonists (42). The scientific evidence suggested that the AKT1 gene modulates both the dopamine release and striatal D2 receptor gene (DRD2) expression in the human brain (43). In our study, we have identified three key targets, all of which belong to G-protein subunits: GNG2, GNG3, and GNG7 (44); Dopamine is reported to control these motor functions, motivation, and some other neuro-regulation through G-protein-coupled receptor (GPCR) signaling (45). Besides, G-protein signaling modulator-3 (GPSM3) could interacts directly with Gα and Gβ subunits of heterotrimeric G proteins to regulate downstream intracellular signals initiated by GPCR that are activated via binding to their cognate ligands. Genome-wide association studies have recently revealed that single nucleotide polymorphisms in GPSM3 are associated with chronic inflammatory diseases (46). A study showed that GHSR-D2R heteromerization could modulate dopamine signaling through a process on G protein conformation (47). Studies showed that estrogen increases the production and release of oxytocin and other neurotransmitters, including dopamine and acetylcholine, which shows the estrogen signaling pathway that may play a significant part in TS (48) are involved in the phosphoinositide 3-kinase (PI3K) kinase and the extracellular signal-regulated kinase (ERK) pathways. Kaempferol was proven to improve motor coordination, raising striatal dopamine and its metabolite levels compared to the MPTP mouse group.
Inflammation and infection
Basal ganglia inflammation and streptococcal infection are assumed to be involved in TS (49); immune responses to β-hemolytic streptococcal infections have triggered many psychiatric diseases, including tic disorders (49-51). Among the critical targets retrieved from the PPI network, TNF gene polymorphisms have been suggested as genetic risk factors for the development of TS (50). A recent study conducted using in vivo TS rat model showed that the inflammatory cytokines IL-6, TNF-α, and IL-1β had an apparent increase in striatum and serum of TS rats (50). Interleukin-8 (CXCL8) is a chemokine family member regulating several tissue functions, including inflammatory conditions, cell activation, and recruitment under homeostatic conditions (52). Research studies found that functional suppression of CXCL8 inhibits neuroinflammation and neuroglial activation via regulating the PI3K/Akt/NF-κB-signaling pathway (53). Signal Transducers and Activators of Transcription (STAT3) play a pivotal role in cell growth and apoptosis by regulating the expression of a range of genes in response to cellular stimuli (54). A study conducted by found STAT3 plays a significant anti-inflammatory role in repairing the damaged tissues (55). The experimental results showed that suppressing the mTOR/STAT3 pathway, LPS-induced apoptosis, and inflammation in rat intestinal epithelial cells was significantly attenuated (56). Either inactivation or hyperactivation of STAT3 may lead to certain diseases or disorders, showing that the tight regulation of STAT3 function is crucial to human well-being (57). It should be noted that hederagenin, one of QD’s hub compounds, could promote the cell apoptosis and inhibit cell proliferation, and these processes were proved to be associated with the inhibition of the STAT3 signaling pathway (58). Epidermal growth factor receptor (EGFR) signaling is regarded as a potential target for many pathological processes and has been proven to be a therapeutic target in infection-induced inflammation (59). The precursor protein proopiomelanocortin (POMC) produces many biologically active peptides via a series of enzymatic steps in a tissue-specific manner, yielding the melanocyte-stimulating hormones (MSHs) (60), and the results of former research suggest that anti-inflammatory influences of neural origin that are triggered by MSH could be used to treat systemic inflammation (61).
MicroRNA (miRNA) pathway
MiRNAs regulate many mRNA expressions, which are small and non-coding RNA molecules. MiRNAs take part in the etiology of various complicated diseases, including several types of cancer, TS, and Fragile X syndrome, etc. (62). The dysregulation of miRNA networks takes part in the development and onset of human neurodegenerative diseases, including TS (63). POMC is a prohormone that encodes multiple smaller peptide hormones within its structure (64); it has been proved leptin can modulate the expression of miRNAs (miR-384-3p, miR-383, and miR-488) that potentially target POMC mRNA (65), which shows POMC probably is a significant node in the miRNA’s process regulation pathway. Oxytocin (Oxt), produced in the supraoptic nuclei and hypothalamic paraventricular, plays a significant role in the central nervous system by influencing diverse behaviors. The study proved that one miRNA, miR-24, to be a novel regulator of Oxt and can regulate both peptide and transcription levels of Oxt (66). Somatostatin (SST) was proven to inhibit the exocrine and endo secretion of this organ (67). Its analogs have shown to regulate the distinct miRNA expression patterns, and both miR-148a and miR-7 display growth-inhibitory attributes (68). Phosphoinositide kinases (PIKs) are a group of lipid kinases that are significant upstream activators of various critical signaling pathways (69). The PIK3CA has been proven to have significant relationships with a range of miRNA regulations (60-71). It has been demonstrated that miRNAs could act an essential role in the EGF triggered signaling cascade and some anti-miRNAs might have considerable effect on this pathway. Simulation results showed that the more targets a miRNA has in the EGFR signaling pathway, the more likely is the corresponding anti-miRNA to exert considerable effect on this pathway (72).
Conclusions
In this study, using the Network Pharmacology, we predicted the genes and protein targets for the QD, which could be involved in QD’s activity in treating TS. The three fundamental processes involved in the observed TS in QD activity are the dopamine system, inflammation and infection, and the miRNA pathway. After analyzing the specific signaling pathways, we have concluded that PI3K-Akt, estrogen, and TNF signaling pathways were three significant pathways involved in the therapeutic effect of QD in TS. The extensive analysis results showed that QD elicits its pharmacological effects in TS by modulating the multiple pathways and multiple targets. Although this study supplies insight into possible molecular pathways involved in QD’s TS activity, further clinical validation studies should confirm the exact molecular mechanism involved.
Acknowledgments
Funding: Grants supported this study from the National Natural Science Foundation of China (No.81774249).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-2158
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-2158). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cavanna AE, Seri S. Tourette’s syndrome. BMJ 2013;347:f4964. [Crossref] [PubMed]
- Muth CC. Tics and Tourette Syndrome. JAMA 2017;317:1592. [Crossref] [PubMed]
- Fan JiFeng. Psychoanalysis and nursing countermeasures of Tic disorder in children. Electronic Journal of Clinical Medical Literature 2010;14:67-68.
- Shapiro AK, Shapiro E, Eisenkraft GJ. Treatment of Gilles de la Tourette’s syndrome with clonidine and neuroleptics. Arch Gen Psychiatry 1983;40:1235-40. [Crossref] [PubMed]
- Cummings DD, Singer HS, Krieger M, et al. Neuropsychiatric effects of guanfa-cine in children with mild tourette syndrome: a pilot study. Clin Neuropharmacol 2002;25:325-32. [Crossref] [PubMed]
- Ross MS, Moldofsky H. A comparison of pimozide and haloperidol in the treat-ment of Gilles de la Tourette’s syndrome. Am J Psychiatry 1978;135:585-7. [Crossref] [PubMed]
- Bruggeman R, Van der Linden C, Buitelaar JK, et al. Risperidone versus pimo-zide in Tourette’s disorder: a comparative double-blind parallel-group study. J Clin Psychiatry 2001;62:50-6. [Crossref] [PubMed]
- Liu ZS, Chen YH, Zhong YQ, et al. A multicenter controlled study on aripiprazole treatment for children with Tourette syndrome in China. Zhonghua Er Ke Za Zhi 2011;49:572-6. [PubMed]
- Wilhelm S, Deckersbach T, Coffey BJ, et al. Habit reversal versus supportive psychotherapy for Tourette’s disorder: a randomized controlled trial. Am J Psychiatry 2003;160:1175-7. [Crossref] [PubMed]
- Song W, Ni S, Fu Y, et al. Uncovering the mechanism of Maxing Ganshi Decoction on asthma from a systematic perspective: A network pharmacology study. Sci Rep 2018;8:17362. [Crossref] [PubMed]
- Qi H, Liu R, Zheng W, et al. Efficacy and safety of traditional Chinese medicine for Tourette’s syndrome: A meta-analysis of randomized controlled trials. Asian J Psychiatr 2020;47:101853. [Crossref] [PubMed]
- Teng QQ, Yan ZJ. Yan Zhaojun treats Tourette syndrome based on Zhiyi dialectical theory. GuoYi Forum 2018;33:23-5.
- Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics 2009;25:2466-72. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Chen CY. TCM Database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One 2011;6:e15939. [Crossref] [PubMed]
- Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13:6964-82. [Crossref] [PubMed]
- Tang F, Tang Q, Tian Y, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Mahuang Fuzi Xixin decoction for application to allergic rhinitis. J Ethnopharmacol 2015;176:402-12. [Crossref] [PubMed]
- Yue SJ, Liu J, Feng WW, et al. System pharmacology-based dissection of the synergistic mechanism of huangqi and huanglian for diabetes mellitus. Front Pharmacol 2017;8:694. [Crossref] [PubMed]
- Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 3: Zooming in on protein-chemical interactions. Nucleic Acids Res 2012;40:D876-80. [Crossref] [PubMed]
- Halladay CW, Trikalinos TA, Schmid IT, et al. Using data sources beyond PubMed has a modest impact on the results of systematic reviews of therapeutic interventions. J Clin Epidemiol 2015;68:1076-84. [Crossref] [PubMed]
- Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014;42:W32-8. [Crossref] [PubMed]
- Xiang SY, Zhao J, Lu Y, et al. Network pharmacology-based identification for therapeutic mechanism of Ling-Gui-Zhu-Gan decoction in the metabolic syndrome induced by antipsychotic drugs. Comput Biol Med 2019;110:1-7. [Crossref] [PubMed]
- Chandran U, Patwardhan B. Network ethnopharmacological evaluation of the immunomodulatory activity of Withania somnifera. J Ethnopharmacol 2017;197:250-56. [Crossref] [PubMed]
- Safran M, Chalifa-Caspi V, Shmueli O, et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res 2003;31:142-6. [Crossref] [PubMed]
- Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res 2014;42:D1091-7. [Crossref] [PubMed]
- Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM ®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789-98. [Crossref] [PubMed]
- Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Missiuro PV, Liu K, Zou L, et al. Information flow analysis of Interactome networks. PLoS Comput Biol 2009;5:e1000350. [Crossref] [PubMed]
- Raman K, Damaraju N, Joshi GK. The organisational structure of protein networks: revisiting the centrality-lethality hypothesis. Syst Synth Biol 2014;8:73-81. [Crossref] [PubMed]
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72. [Crossref] [PubMed]
- Yu G, Zhang Y, Ren W, et al. Network pharmacology-based identification of key pharmacological pathways of Yin-Huang-Qing-Fei capsule acting on chronic bronchitis. Int J Chron Obstruct Pulmon Dis 2016;12:85-94. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2008;4:44-57.
- Chen L, Zhang YH, Wang S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One 2017;12:e0184129. [Crossref] [PubMed]
- Ren G, Zhong Y, Ke G, et al. The Mechanism of Compound Anshen Essential Oil in the Treatment of Insomnia Was Examined by Network Pharmacology. Evid Based Complement Alternat Med 2019;2019:9241403. [Crossref] [PubMed]
- Chen AY, Chen YC. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem 2013;138:2099-107. [Crossref] [PubMed]
- Parveen Z, Deng Y, Saeed MK, et al. Anti-inflammatory and analgesic activities of Thesium Chinense Turcz extracts and its major flavonoids, kaempferol and kaempferol-3-O-glucoside. Yakugaku Zasshi 2007;127:1275-9. [Crossref] [PubMed]
- Lee CW, Park SM, Zhao R, et al. Hederagenin, a major component of Clematis mandshurica Ruprecht root, attenuates inflammatory responses in RAW 264.7 cells and in mice. Int Immunopharmacol 2015;29:528-37. [Crossref] [PubMed]
- Buse J, Schoenefeld K, Münchau A, et al. Neuromodulation in Tourette syndrome: dopamine and beyond. Neurosci Biobehav Rev 2013;37:1069-84. [Crossref] [PubMed]
- Maia TV, Conceição VA. Dopaminergic Disturbances in Tourette Syndrome: An Integrative Account. Biol Psychiatry 2018;84:332-44. [Crossref] [PubMed]
- Nash AI. Crosstalk between insulin and dopamine signaling: A basis for the metabolic effects of antipsychotic drugs. J Chem Neuroanat 2017;83-84:59-68. [Crossref] [PubMed]
- Cai W, Xue C, Sakaguchi M, et al. Insulin regulates astrocyte gliotransmission and modulates behavior. J Clin Invest 2018;128:2914-26. [Crossref] [PubMed]
- Diaz-Ruiz O, Zapata A, Shan L, et al. Selective deletion of PTEN in dopamine neurons leads to trophic effects and adaptation of striatal medium spiny projecting neurons. PLoS One 2009;4:e7027. [Crossref] [PubMed]
- Shumay E, Wiers CE, Shokri-Kojori E, et al. New Repeat Polymorphism in the AKT1 Gene Predicts Striatal Dopamine D2/D3 Receptor Availability and Stimulant-Induced Dopamine Release in the Healthy Human Brain. J Neurosci 2017;37:4982-91. [Crossref] [PubMed]
- Syrovatkina V, Alegre KO, Dey R, et al. Regulation, Signaling, and Physiological Functions of G-Proteins. J Mol Biol 2016;428:3850-68. [Crossref] [PubMed]
- Billard MJ, Gall BJ, Richards KL, et al. G protein signaling modulator-3: a leukocyte regulator of inflammation in health and disease. Am J Clin Exp Immunol 2014;3:97-106. [PubMed]
- Liu C, Kaeser PS. Mechanisms and regulation of dopamine release. Curr Opin Neurobiol 2019;57:46-53. [Crossref] [PubMed]
- Damian M, Pons V, Renault P, et al. GHSR-D2R heteromerization modulates dopamine signaling through an effect on G protein conformation. Proc Natl Acad Sci USA 2018;115:4501-6. [Crossref] [PubMed]
- Paletta P, Sheppard PAS, Matta R, et al. Rapid effects of estrogens on short-term memory: Possible mechanisms. Horm Behav 2018;104:88-99. [Crossref] [PubMed]
- Madhusudan N, Cavanna AE. The role of immune dysfunctionin the development of tics and susceptibility to infections in Tourette syndrome: a systematic review. Basal Ganglia 2013;3:77-84. [Crossref]
- Kansy JW, Katsovich L, Bibb JA, et al. Identification of pyruvate kinase asan antigen associated with Tourette syndrome. J Neuroimmunol 2006;181:165-76. [Crossref] [PubMed]
- Keszler G, Kruk E, Kenezloi E, et al. Association of the tumor necrosis factor -308 A/G promoter polymorphism with Tourette syndrome. Int J Immunogenet 2014;41:493-8. [Crossref] [PubMed]
- Hongyan L, Mengjiao Z, Chunyan W, et al. Rhynchophyllin attenuates neuroinflammation in Tourette syndrome rats via JAK2/STAT3 and NF-κB pathways. Environ Toxicol 2019;34:1114-20. [Crossref] [PubMed]
- Russo RC, Garcia CC, Teixeira MM, et al. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev Clin Immunol 2014;10:593-619. [Crossref] [PubMed]
- Fan Y, Mao R, Yang J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013;4:176-85. [Crossref] [PubMed]
- Hu YS, Han X, Liu XH. STAT3: A Potential Drug Target for Tumor and Inflammation. Curr Top Med Chem 2019;19:1305-17. [Crossref] [PubMed]
- Li L, Wan G, Han B, et al. Echinacoside alleviated LPS-induced cell apoptosis and inflammation in rat intestine epithelial cells by inhibiting the mTOR/STAT3 pathway. Biomed Pharmacother 2018;104:622-8. [Crossref] [PubMed]
- Hillmer EJ, Zhang H, Li HS, et al. STAT3 signaling in immunity. Cytokine Growth Factor Rev 2016;31:1-15. [Crossref] [PubMed]
- Fang L, Liu M, Cai L. EGFR in infection-induced inflammation. Chinese Journal of Cellular and Molecular Immunology 2019;35:140-5. [PubMed]
- Millington GW. Proopiomelanocortin (POMC): the cutaneous roles of its melanocortin products and receptors. Clin Exp Dermatol 2006;31:407-12. [Crossref] [PubMed]
- Catania A, Delgado R, Airaghi L, et al. Alpha-MSH in systemic inflammation. Central and peripheral actions. Ann N Y Acad Sci 1999;885:183-7. [Crossref] [PubMed]
- Hardbower DM, Singh K, Asim M, et al. EGFR regulates macrophage activation and function in bacterial infection. J Clin Invest 2016;126:3296-312. [Crossref] [PubMed]
- Perkins DO, Jeffries CD, Jarskog LF, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 2007;8:R27. [Crossref] [PubMed]
- Ha TY. MicroRNAs in Human Diseases: From Autoimmune Diseases to Skin, Psychiatric and Neurodegenerative Diseases. Immune Netw 2011;11:227-44. [Crossref] [PubMed]
- Cawley NX, Li Z, Loh YP. 60 YEARS OF POMC: Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. J Mol Endocrinol 2016;56:T77-T97. [Crossref] [PubMed]
- Derghal A, Astier J, Sicard F, et al. Leptin Modulates the Expression of miRNAs-Targeting POMC mRNA by the JAK2-STAT3 and PI3K-Akt Pathways. J Clin Med 2019;8:2213. [Crossref] [PubMed]
- Sliwińska-Mossoń M, Veselý M, Milnerowicz H. The clinical significance of somatostatin in pancreatic diseases. Ann Endocrinol (Paris) 2014;75:232-40. [Crossref] [PubMed]
- Døssing KBV, Kjær C, Vikeså J, et al. Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells. Genes (Basel) 2018;9:337. [Crossref] [PubMed]
- Alqahtani A, Ayesh HSK, Halawani H. PIK3CA Gene Mutations in Solid Malignancies: Association with Clinicopathological Parameters and Prognosis. Cancers (Basel) 2019;12:93. [Crossref] [PubMed]
- Baldassari F, Zerbinati C, Galasso M, et al. Screen for MicroRNA and Drug Interactions in Breast Cancer Cell Lines Points to miR-126 as a Modulator of CDK4/6 and PIK3CA Inhibitors. Front Genet. 2018;9:174. [Crossref] [PubMed]
- Guo Y, Lang X, Lu Z, et al. MiR-10b Directly Targets ZEB1 and PIK3CA to Curb Adenomyotic Epithelial Cell Invasiveness via Upregulation of E-Cadherin and Inhibition of Akt Phosphorylation. Cell Physiol Biochem 2015;35:2169-80. [Crossref] [PubMed]
- Li J, Pandey V, Kessler T, et al. Modeling of miRNA and drug action in the EGFR signaling pathway. PLoS One 2012;7:e30140. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)