Drug discovery in cardiovascular disease identified by text mining and data analysis
Introduction
According to the World Health Organization (WHO) health report, atherosclerotic cardiovascular disease (ASCVD) is one of the most common cardiovascular diseases and one of the leading causes to death around the world (1). In 2012, the death toll of the cardiovascular disease reached 17.5 million, accounting for more than 31% of the global deaths. ASCVD is a chronic disease, which has two primary forms: one is ischaemic heart disease; another is a cerebrovascular disease (2). The occurrence of ASCVD has multiple intrinsic factors, which include the accumulation of immune cells (such as microphages), production of pro-inflammatory cytokines (CCL2, TNFα, IL-1β, and IL-6) and dysfunction of endothelial and vascular smooth muscle cells (3-5). The external risk factors of cardiovascular disease include hypertension, hyperlipidemia, high cholesterol, diabetes, obesity, smoking, and an unhealthy lifestyle. Cardiovascular complications are a major threat to patients with type II diabetes and new updated strategies for prevention are urgently demanded (6). Current available myriad studies have revealed that atherosclerosis could occur in any arterial blood vessels, and lead to many related diseases, such as ischemic heart, peripheral artery and chronic kidney diseases, etc. (7-12).
With the development of bioinformatics, text mining and data analysis have been used in many aspects, like as the identification of potential key gene targets, signal pathway, diagnosis of diseases and prediction of the occurrence of diseases. Meanwhile, researchers have discovered a series of mechanisms of atherosclerosis by doing experiments (13). Compared with bioinformatics in the cancer field, there are fewer researches focused on cardiovascular diseases through text mining and data analysis.
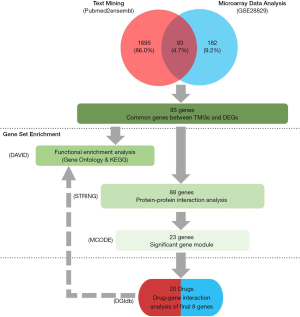
In this study, we first used several bioinformatics strategies, such as text mining and microarray data analysis, to obtain the common genes, which their gene expressions have changed between advanced atherosclerosis plaque and early atherosclerosis plaque group, and the correlation between/within genes have existed in these genes. Then we clustered these genes in protein and protein interactions (PPI) and identified significant module genes that have more interactions. Lastly, drug-gene interaction of module genes was carried out in the drug gene interaction database (DGIdb), and aimed at finding some existed drugs to provide new ideas and basis for the prevention and treatment of cardiovascular disease. Figure 1 shows the framework of this study.
Methods
Text mining, microarray data analysis
Firstly, the open-access website pubmed2ensembl was used to perform text mining (http://pubmed2ensembl.ls.manchester.ac.uk). When a keyword is entered, the pubmed2ensembl website can retrieve and extract all gene symbols found in published PubMed articles related to keyword (14). We input the “Atherosclerosis” keyword into pubmed2ensembl and then extracted all unduplicated genes, these gene sets make up the text mining genes (TMGs).
Secondly, GSE28829 raw expression profiles (.CEL format files) and meta clinical information data (.soft format file) have been retrieved and downloaded from NCBI-GEO website (15,16), which was executed on the GPL570 platform. GSE28829 datasets contain 16 advanced atherosclerosis plaques (EA) and 13 early atherosclerosis plaques (AA) from carotid artery samples (17,18). We transformed probe identification numbers into official gene symbols via the information built in GPL570 platform, the non-mRNA probes were removed and the multiple probes to the same gene were deal with to retain the significant value as the gene expression level. Then, we used affy, affyPLM, and limma packages loaded on R/Rstudio to detect the differentially expressed genes (DEGs) between EA and AA samples (19,20). DEGs with the |log2 fold change (FC)| ≥1 and adjust P value <0.05, corrected by the BH method, as the threshold criteria were selected for the follow-up analyses.
The intersection of TMGs and DEGs were overlapping genes, and then for the next further analysis.
Gene ontology (GO) and pathway enrichment analysis
The GO analysis is a ubiquitous and useful method for annotating gene products and its features of functional characteristics. GO is categorized into three groups (biological process, cellular component, molecular function). The Kyoto Encyclopedia of Genes and Genomes (KEGG) (21) is an open access informatic database from Japan for interpreting biological function and characteristics of the organic system, generated by the gene-chip and high-throughput experiments. The GO and KEGG enrichment of Overlapping genes were analyzed via an online tool DAVID, a functional annotation bioinformatics microarray analysis website, used to gene annotation, visualization. P<0.05 was considered as statistically significant as the threshold.
Protein interaction and module analysis
STRING database (version 11.0), covering about 24.6M proteins and more than 3.1 billion interactions originated from 5.09K organisms, was known as the primary source to depict and display the interaction among various proteins. Firstly, we uploaded overlapping genes into the STRING website, and a minimum interaction score >0.4 (low confidence) was set as significant threshold. Then the TSV format file of protein-protein interaction (PPI) was downloaded, and PPI networks were constructed by Cytoscape software. Subsequently, the Molecular Complex Detection (MCODE) and STRING app built in Cytoscape was utilized to classify the significant gene modules (clusters), which have highly interconnected clusters in the PPI network. The parameters in MCODE were set by default. The genes in gene modules were performed drug-gene interaction analysis.
Drug-gene interaction and functional analysis of potential genes
The drug-gene interaction database (DGIdb: http://www.dgidb.org) is an open-source and supports searching, browsing and filtering of information on drug-gene interactions based on over thirty trusted sources. The module genes, as the potential targets, were pasted into the drug-gene database to search for existing drugs or compounds. These potential genes which have matched drugs were obtained and also performed functional enrichment analysis.
Statistics analysis
The moderate t-test was applied to identify DEGs, Fisher’s Exact test was used to analyzed GO and KEGG enrichments (22). All statistical analysis was executed in R version 3.6.1 software.
Results
TMGs & DEGs acquisition
Based on the text mining and microarray data analysis strategy that is described in the methods section, 1,788 TMGs were related to atherosclerosis, 275 DEGs were identified in AA compared with EA group, and 93 genes were overlapping between TMGs and DEGs (Figure 1). Among these overlapping genes, 89 genes were up-regulated genes and 4 genes were down-regulated genes (Table 1).

Full table
GO and pathway enrichment analysis
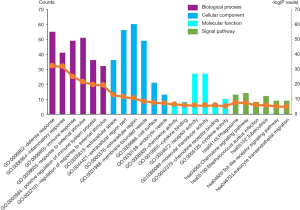
In order to demonstrate GO and signal pathway enrichments of common genes, we executed functional annotation in DAVID website. As shown in Figure 2, it showed that the top six significant enrichment terms for BP, CC, MF, and KEGG signal pathways of common genes. In BP category, it was mainly enriched in the defense response, inflammatory response, and response to external stimulus. In CC category, it was significantly enriched in the extracellular space, extracellular region part, and extracellular part. In MF category, it was mainly enriched in the chemokine activity, cytokine binding, and receptor activity. As for signal pathway enrichment, it was mainly involved in the phagosome, chemokine signaling pathway, and staphylococcus aureus infection terms, respectively (Figure 2).
Protein interaction and module analysis
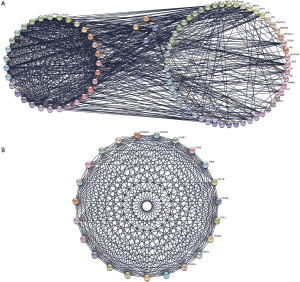
All of the common genes were pasted into the STRING website and then analyzed with Cytoscape software. A total of 88 genes/nodes with 705 edges were involved in the construction of the PPI networks, and 5 genes didn’t fall into the PPI networks (Figure 3A). To get the most significant gene modules, a significant gene module was selected by using the MCODE application. Module 1 consists 23 genes/nodes with 232 edges/interactions, which all exhibit up-regulated (Figure 3B).
Drug-gene interaction and functional analysis of potential genes
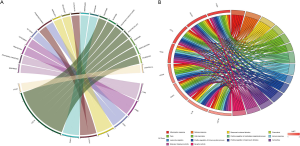
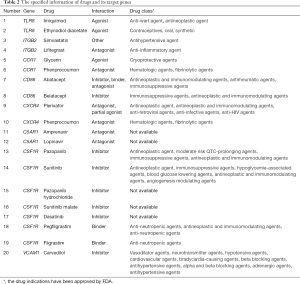
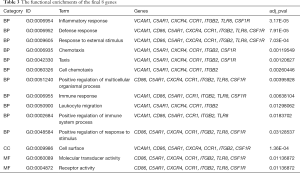
The 23 genes clustered in the significant gene module were selected for drug-gene interaction analysis. We found that there were eight genes target to 20 potential existing drugs, which have been divided into 4 drug-gene interaction types and all have their initial drug indications (Figure 4A, Table 2). Besides, the functional enrichment terms of those eight gene were mainly involved in 11 GO terms (Figure 4B, Table 3), the most significant GO terms were the inflammatory response (BP, adj.pval =3.17E-04), cell surface (CC, adj.pval =1.36E-04), molecular transducer activity (MF, adj.pval =1.13E-02), respectively (Figure 4B, Table 3).
Full table
Full table
Availability of data and materials
The data associated with this article has been deposited in the NCBI-GEO website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Discussion
Atherosclerotic cardiovascular disease is considered to be a common and chronic inflammatory malady of the arterial intima. In this study, our purposes were to discover new drug indications. As we expected, we finally have found eight genes, namely VCAM1, CSF1R, C5AR1, CXCR4, CD86, CCR1, ITGB2, and TLR8, target to 20 existing potential drugs which might be used for the treatment of atherosclerosis-related diseases.
In our study, we have identified 20 drugs, which have target to eight genes (VCAM1, CSF1R, C5AR1, CXCR4, CD86, CCR1, ITGB2, and TLR8). These genes all have correlation with atherosclerosis. Almeda et al. team have discovered that the vascular cell adhesion molecule-1 (VCAM1) exposed to cytokines have shown potential for drug delivery (23). The inhibition of the progression of atherosclerosis has related to the interaction between miR-155 and B-cell leukemia/lymphoma, when miR-155 suppressed macrophage proliferation by targeting colony-stimulating factor-1 receptor (CSF1R) (24). High levels of C5a receptor 1 (C5AR1) in human lung tumors were significantly associated with overall survival and bone metastasis (25). Type 4 chemokine receptor (CXCR4) participated in immune cell migration during the progression of atherosclerosis (26) and its expression level has been treated as a promising target for molecular imaging in atherosclerosis (27). CD86 is expressed on the antigen-presenting cells (APCs) and provides costimulatory signals to T cells. Meletta et al. adopt CD86/CD80 as an imaging probe for atherosclerosis diagnosis (28). Han et al. have demonstrated that CC chemokine ligand 7 (CCL7) and its receptors CCR1, CCR2, and CCR3 were high expressions significantly in lung cancer bone metastasis (29). Moreover, CCR1 is also a target of the EGF-AKT-mTOR-STAT3 signaling pathway in breast cancer (30). The differentially methylated regions in integrin subunit beta 2 (ITGB2) may mediate genetic risk in inflammatory disease (31).
Toll-like receptor 8 (TLR8), a member of TLRs, has correlation with inflammatory disease (32).
A large amount of recent evidence confirms that the occurrence and development of atherosclerotic cardiovascular disease is a chronic inflammatory process. Inflammation is one of the most important intrinsic factors that cause plaque vulnerability. Based on previous scientific research, Janus-activated kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway play an important role in inflammation, chronic pain, and even have an effect on the pathogenesis of neuropathic pain. Proinflammatory cytokines interleukin (IL-1β) is a downstream factor involved in JAK2/STAT3 signal transition pathway, which participates in neuron injury and inflammation (33). As the important signal pathway connecting inflammation and tumor, activated signal transduction and transcriptional activation factor 3 (STAT3) leads to genetic abnormal expression, gene silencing, genomic instability, etc. in non-small cell lung cancer (NSCLC) cells, and induces therapeutic resistance (34).
Currently, the interaction between the drugs and genes we discovered were divided into four types, namely agonist, inhibitor, antagonist, and binder (Table 3). These drugs are classified into anti-inflammatory agent, anti-neoplastic and immunomodulating agents. Although these existing drugs provide a new perspective for us to study cardiovascular diseases, further clinical trials need to be performed for confirmation of its new function and indications.
Conclusions
According to the text mining conception (keyword: atherosclerosis) and microarray data analysis (dataset: GSE28829), we found 20 existing drugs, approved by FDA, target to eight genes, which involved in the inflammatory pathway. These genes might be used for atherosclerosis, as well as its original drug indications.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81770294).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-705). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. For human datasets mentioned in this study, please refer to the original article (PMID: 22388324). We just re-analyzed the open accessed datasets, and no ethical approval was required by the local ethics committees.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Carrizales-Sepulveda EF, Ordaz-Farias A, Vera-Pineda R, et al. Periodontal Disease, Systemic Inflammation and the Risk of Cardiovascular Disease. Heart Lung Circ 2018;27:1327-34. [Crossref] [PubMed]
- Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation 2017;135:e146-603. [Crossref] [PubMed]
- Ridker PM, Luscher TF. Anti-inflammatory therapies for cardiovascular disease. Eur Heart J 2014;35:1782-91. [Crossref] [PubMed]
- Khambhati J, Engels M, Allard-Ratick M, et al. Immunotherapy for the prevention of atherosclerotic cardiovascular disease: Promise and possibilities. Atherosclerosis 2018;276:1-9. [Crossref] [PubMed]
- Cui C, Wang X, Shang XM, et al. lncRNA 430945 promotes the proliferation and migration of vascular smooth muscle cells via the ROR2/RhoA signaling pathway in atherosclerosis. Mol Med Rep 2019;19:4663-72. [Crossref] [PubMed]
- Zweck E, Roden M. GLP-1 receptor agonists and cardiovascular disease: drug-specific or class effects? Lancet Diabetes Endocrinol 2019;7:89-90. [Crossref] [PubMed]
- Aboyans V, Ricco JB, Bartelink MLE, et al. 2017 ESC Guidelines on the Diagnosis and Treatment of Peripheral Arterial Diseases, in collaboration with the European Society for Vascular Surgery (ESVS): Document covering atherosclerotic disease of extracranial carotid and vertebral, mesenteric, renal, upper and lower extremity arteries Endorsed by: the European Stroke Organization (ESO)The Task Force for the Diagnosis and Treatment of Peripheral Arterial Diseases of the European Society of Cardiology (ESC) and of the European Society for Vascular Surgery (ESVS). Eur Heart J 2018;39:763-816. [Crossref] [PubMed]
- Levey AS, Eckardt KU, Tsukamoto Y, et al. Definition and classification of chronic kidney disease: a position statement from Kidney Disease: Improving Global Outcomes (KDIGO). Kidney Int 2005;67:2089-100. [Crossref] [PubMed]
- Anand SS, Yusuf S, Vuksan V, et al. Differences in risk factors, atherosclerosis, and cardiovascular disease between ethnic groups in Canada: the Study of Health Assessment and Risk in Ethnic groups (SHARE). Lancet 2000;356:279-84. [Crossref] [PubMed]
- Criqui MH, Aboyans V. Epidemiology of peripheral artery disease. Circ Res 2015;116:1509-26. [Crossref] [PubMed]
- DeFronzo RA, Ferrannini E. Insulin Resistance: A Multifaceted Syndrome Responsible for NIDDM, Obesity, Hypertension, Dyslipidemia, and Atherosclerotic Cardiovascular Disease. Diabetes Care 1991;14:173-94. [Crossref] [PubMed]
- Kullo IJ, Rooke TW. Peripheral artery disease. N Engl J Med 2016;374:861-71. [Crossref] [PubMed]
- Yuan Y, Long P, Liu K, et al. Multiple plasma metals, genetic risk and serum C-reactive protein: A metal-metal and gene-metal interaction study. Redox Biol 2020;29:101404. [Crossref] [PubMed]
- Baran J, Gerner M, Haeussler M, et al. pubmed2ensembl: a resource for mining the biological literature on genes. PLoS One 2011;6:e24716. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37:D885-90. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res 2007;35:D760-5. [Crossref] [PubMed]
- Döring Y, Manthey HD, Drechsler M, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation 2012;125:1673-83. [Crossref] [PubMed]
- Larriba Y, Rueda C, Fernandez MA, et al. Microarray Data Normalization and Robust Detection of Rhythmic Features. Methods Mol Biol 2019;1986:207-25. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc 1992;85:87-94. [Crossref]
- Almeda D, Wang B, Auguste DT. Minimizing antibody surface density on liposomes while sustaining cytokine-activated EC targeting. Biomaterials 2015;41:37-44. [Crossref] [PubMed]
- Wei Y, Zhu M, Corbalán-Campos J, et al. Regulation of Csf1r and Bcl6 in macrophages mediates the stage-specific effects of microRNA-155 on atherosclerosis. Arterioscler Thromb Vasc Biol 2015;35:796-803. [Crossref] [PubMed]
- Ajona D, Zandueta C, Corrales L, et al. Blockade of the complement C5a/C5aR1 axis impairs lung cancer bone metastasis by CXCL16-mediated effects. Am J Respir Crit Care Med 2018;197:1164-76. [Crossref] [PubMed]
- Li X, Yu W, Wollenweber T, et al. [68 Ga] Pentixafor PET/MR imaging of chemokine receptor 4 expression in the human carotid artery. Eur J Nucl Med Mol Imaging 2019;46:1616-25. [Crossref] [PubMed]
- Weiberg D, Thackeray JT, Daum G, et al. Clinical molecular imaging of chemokine receptor CXCR4 expression in atherosclerotic plaque using 68Ga-pentixafor PET: correlation with cardiovascular risk factors and calcified plaque burden. J Nucl Med 2018;59:266-72. [Crossref] [PubMed]
- Meletta R, Herde AM, Dennler P, et al. Preclinical imaging of the co-stimulatory molecules CD80 and CD86 with indium-111-labeled belatacept in atherosclerosis. EJNMMI Res 2016;6:1. [Crossref] [PubMed]
- Han S, Wang T, Chen Y, et al. High CCL7 expression is associated with migration, invasion and bone metastasis of non-small cell lung cancer cells. Am J Transl Res 2019;11:442-52. [PubMed]
- Shin SY, Da Hyun Lee JL, Choi C, et al. CC motif chemokine receptor 1 (CCR1) is a target of the EGF-AKT-mTOR-STAT3 signaling axis in breast cancer cells. Oncotarget 2017;8:94591. [Crossref] [PubMed]
- Ventham N, Kennedy NA, Adams A, et al. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun 2016;7:13507. [Crossref] [PubMed]
- Kapelouzou A, Giaglis S, Peroulis M, et al. Overexpression of Toll-Like Receptors 2, 3, 4, and 8 is correlated to the vascular atherosclerotic process in the hyperlipidemic rabbit model: The effect of statin treatment. J Vasc Res 2017;54:156-69. [Crossref] [PubMed]
- Xu M, Ni H, Xu L, et al. B14 ameliorates bone cancer pain through downregulating spinal interleukin-1β via suppressing neuron JAK2/STAT3 pathway. Mol Pain 2019;15:1744806919886498. [Crossref] [PubMed]
- Sun S, Jin S, Guo R. Role of STAT3 in Resistance of Non-small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi 2019;22:457-63. [PubMed]