
mRNA expression disturbance of complement system related genes in acute arterial thrombotic and paroxysmal atrial fibrillation patients
Introduction
Coronary atherosclerosis and plaque rupture, which would result in acute arterial thrombotic events, are main pathologic changes in patients with acute myocardial infarction (AMI) (1). Innate immunity, including the complement system, is important in the formation of atherosclerotic plaques (2). Paroxysmal atrial fibrillation (PAF) is a common arrhythmia, which often causes thrombi in left atrium. The treatment of PAF has changed from an antiplatelet therapy into an anticoagulant therapy (3). Though it has been established that atrial fibrillation is associated with the increased inflammatory activity (4), there are few studies on atrial fibrillation and complement system.
The formation of arterial and venous thrombi is associated with the significant decrease or disturbance of immune cell balancing function (5,6). The complement system not only belongs to the innate immune system, but also its components act as cofactors or enhancement factors of antibodies in adaptive immune responses. It has been demonstrated that pathogenesis of AMI was related to inflammations. Previous studies have shown that the complement system play a major role in the generation of AMI related inflammatory responses (7,8). In order to identify the immune functions in AMI and PAF patients, there is a need to test every protein in the complement system, where difficulties exist due to the various kinds of proteins. Thus, this study was designed to test the gene expression related to innate components, receptors and regulatory proteins of the complement system in peripheral blood mononuclear cells from the AMI group, the PAF group and the controls [stable angina pectoris (SAP) patients] with human genomics technology.
Methods
Patients
Twenty patients (18 males and 2 females, average age: 57.8±11.9 years) admitted in Tongji Hospital due to AMI during 2018 January and July were enrolled into the AMI group. All these patients were admitted into Cardiac Care Unit within 12 hours after symptoms appeared. Diagnostic criteria of AMI: elevated serum myocardium marker (troponin) to at least 99% upper limit of reference value and one of the following clinical tests: (I) ischemic symptoms; (II) novel ischemic electrocardiograph changes (new ST-T change or left bundle branch block); (III) formation of pathological Q wave in ECG; (IV) image evidences showed activities loss of myocardium or abnormal activities of local ventricular wall; (V) coronary arteriography showed coronary arterial thrombus. PAF and other concomitant diseases were excluded.
Another 20 novel PAF patients (18 males and 2 females, average age: 65.4±12.5 years) were recruited into the PAF group during the same period. The diagnosis of PAF based on the results of electrocardiogram, which should indicate a presence of continuous and irregular atrial fibrillation waves instead of P wares. AMI, SAP and other concomitant diseases were excluded.
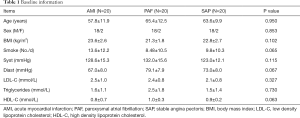
Another 20 SAP patients (18 males and 2 females, average age: 63.6±9.9 years) were recruited into the control group during the same period. The criteria of SAP were as follows: angina pectoris of effort, positive treadmill test and 70% stenosis of at least one coronary artery in coronary arteriography. AMI, PAF and other concomitant diseases were excluded. There were no significant differences of age, gender, smoke, body mass index, systolic blood pressure, diastolic blood pressure, low density lipoprotein, high density lipoprotein and triglyceride among three groups (P>0.05) (Table 1). This study has been approved by the Ethics Committee of Tongji Hospital (No. KYSB-2014-054), and informed consent form was also obtained. This study has been performed in accordance with the principles of Declaration of Helsinki.
Full table
Total RNA isolation
A total of 5 mL of peripheral blood samples anti-coagulated with EDTA were drawn from AMI and SA patients, immediately after being admitted to the hospital. And also 5 mL peripheral blood samples were drawn from the controls. Leucocytes were obtained through density gradient centrifugation with Ficoll solution and the remaining red blood cells were destroyed by erythrocyte lysis buffer (Qiagen, Hilden, Germany). Total mononuclear cell RNA was extracted with TRIzol (Invitrogen, Carlsbad, USA) and purified with QiagenRNeasy column (Qiagen), according to the manufacturer’s instructions. The isolated total RNA was tested and quantified using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technology, Cambridge, UK).
Gene expression clip
Agilent G4112A Whole Human Genome Oligo Microarrays were purchased from Agilent (USA). There are 314 negative control spots, 1,924 positive control spots and 359 blank spots. The functions of more than 70% of the genes in the microarray are already known. All objects were subjected to clip analysis.
Target preparation and microarray hybridization
The RNA samples of three groups were labeled using the indirect labeling method. Briefly, 1 µg of total RNA was reverse transcribed. Second strand cDNA was then produced and purified followed by in vitro transcription with T7 RNA Polymerase. During in vitro transcription, the modified nucleotide, 5-(3-aminoallyl)-UTP (aaUTP) was incorporated into the cDNA. Subsequently, the fluorescent Cy3 was chemically coupled with the aaUTP which contains a reactive primary amino group on the C5 position of uracil. The dye incorporation rate was assessed with a Nanodrop ND-1000 spectrophotometer and was found to be between 1.2 1.4 pmol/µL. Hybridization was carried out using the Agilent Oligonucleotide Microarray in situ Hybridization Plus kit (p/n 5,184-3,568), according to the manufacturer’s instructions. Briefly, 750 ng of Cy3-labeled sample cDNA was subjected to fragmentation (30 min at 60 °C) and then hybridization on 44K Human Whole-Genome 60-mer oligo-chips (G4112F, Agilent Technologies) in a rotary oven (10 rpm, 60 °C, 17 h). Slides were disassembled and washed in solutions I and II, according to the manufacturer’s instructions.
RT-PCR
Three differential genes were selected and their expressions were confirmed by RT-PCR. Among the genes with differential expressions, 7 genes were randomly selected and the house keeping genes GAPDH were subjected to RT-PCR. The relative expression levels were indicated as the expression of the target genes normalized to the expression of GAPDH (2-ΔΔCt). The melting curve and the 2-ΔΔCt-method were used to compare the differences in the expressions between the control and the PE group. The results from RT-PCR were consistent with from the microarray analysis.
Statistical analysis
Measurement data were expressed by average ± standard deviation. The Agilent Feature Extraction software was used to collect the original data from the microarray, followed by an analysis with a robust multichip average. The gene intensity data between the PE and control group were compared with a random variance model-corrected t-test by SPSS 14.0 software packet. Differentially expressed genes were identified from whole genomes. P<0.05 was considered statistically significant.
Results
mRNA expression of genes related to complement recognition and activation
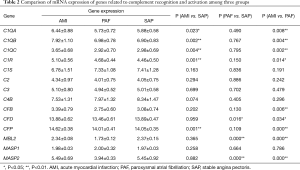
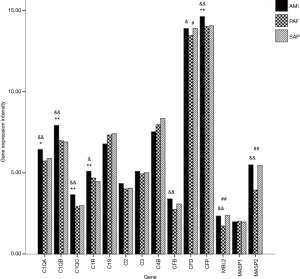
A total of 14 related genes were detected. When compared with the control group, mRNA expression of C1QA, C1QB, C1QC, C1R and CFP genes was significantly upregulated in AMI patients (P<0.05); and mRNA expression of CFD, MBL2 and MASP2 genes was significantly downregulated in PAF patients (P<0.05). Results of the comparison between the AMI and PAF group showed that mRNA expression of C1QA, C1QB, C1QC, C1R, CFB, CFD, CFP, MBL2 and MASP2 genes was significantly upregulated in the AMI group (P<0.05) (Table 2 and Figure 1).
Full table
mRNA expression of genes related to membrane attack complex (MAC)
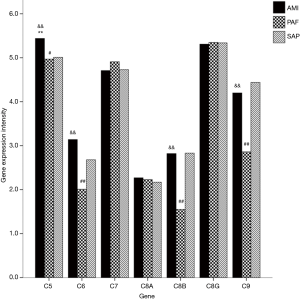
A total of seven genes related to MAC were detected. When compared with the controls, mRNA expression of C5 gene was significantly upregulated in AMI patients (P<0.05); and mRNA expression of C5, C6, C8B and C9 genes was significantly downregulated in PAF group (P<0.05). When compared with the PAF group, mRNA expression of C5, C6, C8B and C9 genes was significantly upregulated in AMI patients (P<0.05) (Table 3 and Figure 2).

Full table
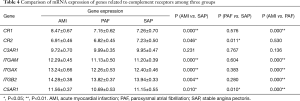
mRNA expression of genes related to complement receptors
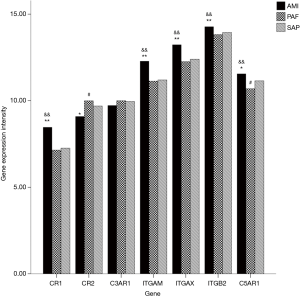
A total of seven genes related to complement receptors were detected. When compared with the control group, mRNA expression of CR1, ITGAM, ITGAX, ITGB2 and C5AR1 genes was significantly upregulated, and CR2 gene significantly downregulated in the AMI group (P<0.05); and mRNA expression of C5AR1 and CR2 genes was significantly downregulated in PAF group (P<0.05). When compared with the PAF group, mRNA expression of CR1, ITGAM, ITGAX, ITGB2 and C5AR1 genes was significantly upregulated in AMI patients (P<0.05) (Table 4 and Figure 3).
Full table
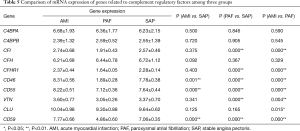
mRNA expression of genes related to complement regulatory factors
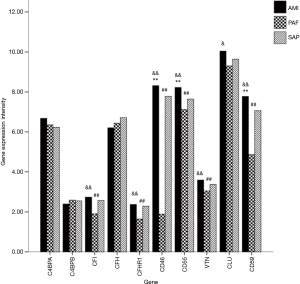
A total of 10 genes related to regulatory factors in the complete system were detected. When compared with the control group, mRNA expression of CD46, CD55 and CD59 genes was significantly upregulated in AMI group; and mRNA expression of CFI, CFHR1, CD46, CD55, VTN and CD59 genes was significantly downregulated in PAF group (P<0.05). When compared with the PAF group, mRNA expression of CFI, CFHR1, CD46, CD55, CLU, VTN and CD59 genes was significantly upregulated in AMI group (P<0.05) (Table 5 and Figure 4).
Full table
Discussion
The complement is not a single molecule, but a group of more than 30 kinds of soluble and membrane-bound proteins with enzymatic properties existing in serum, tissue fluid, and cell membrane, which could be activated by a series of cascade reactions. Taken together, they are called the complement system. Its main functions consist of mediation of inflammations, opsonized phagocytosis, and lysis of cell membranes. In this study, a total mRNA expression of 38 genes was detected and analyzed in four aspects of the complement system, which are early components, late components, complement receptors, and complement regulatory factors.
Through three pathways, including classical pathway, alternative pathway and lectin pathway, can the complement cascade be activated. As the initial part of classical pathway, C1 is a compound of C1q, C1r and C1s. C1q serves as a subunit with a recognition function, while C1r and C1s act as catalytic subunits. C1q is a protein formed by A, B and C peptide chains, which are encoded by C1QA, C1QB and C1QC genes respectively. And all these chains spin and fold to generate different active sites, which can bind with C1R gene encoded C1r and C1S gene encoded C1s. C1q can be activated by combining with an IgM or two close IgG, meanwhile the inhibitory function of C1INH on C1q will be removed. Besides, it was found that C1q could stimulate B lymphocytes to produce more immunoglobulins to combine with, which could increase the possibility of combination with IgM and IgG. In this study, mRNA expression of genes related to C1, representing the activation of classical pathway, was significantly upregulated in AMI patients when compared with the controls, indicating that the classical pathway was activated in AMI patients. Other researchers also confirmed the classic pathway activation in acute coronary syndrome (9,10). However, there was no significant difference between PAF patients and the controls, suggesting that the classical pathway was not activated in PAF patients. As for the comparison between PAF and AMI patients, it was without doubt that mRNA expression of C1QA, C1QB, C1QC and C1R genes in AMI patients was significantly upregulated, indicating the activation of the classical pathway was significantly stronger in AMI patients than in PAF patients.
C3 locates at the meeting point of three activation pathways, and thus plays a pivotal role in the whole activation process of the complement system. C3 can be hydrolyzed into C3a and C3b. The activation of alternative pathway is dependent on C3. The B and D factors, encoded by CFB and CFD respectively, are involved in activating C3b into C3 convertase (11); and the P factor, encoded by CFP, can positively regulates C3 convertase to accelerate the alternative pathway. Studies showed that C3 activation was associated with increased risks in coronary heart diseases (12,13). In this study, mRNA expression of CFP gene was significantly upregulated in AMI patients, indicating that the C3 convertase of alternative pathway in AMI patients was positively and significantly regulated. However, mRNA expression of CFD gene was significantly downregulated in PAF patients when compared with the controls, suggesting that the formation of C3 convertase in the alternative pathway was inhibited in PAF patients. As for the comparison between AMI and PAF patients, the significantly upregulated mRNA expression of CFB, CFD and CFP genes in AMI patients indicated that the activation of C3 convertase in the alternative pathway in AMI patients was significantly more than that in PAF patients.
As for the lectin pathway, it is activated through proteins encoded by MBL, MASP1 and MASP2 genes, which afterwards activate C4 directly. When compared with the control group in this study, there was no significant change of mRNA expression of MBL, MASP1 and MASP2 genes in the AMI group, suggesting that the lectin pathway was not significantly activated in AMI patients. It was also reported that some variants in genes encoding MBL are associated with risks for coronary heart diseases (14). However, mRNA expression of MBL2 and MASP2 genes was significantly downregulated in PAF patients, indicating that the lectin pathway was significantly inhibited in PAF patients. When the AMI patients was compared with the PAF patients, mRNA expression of MBL2 and MASP2 genes was significantly upregulated in AMI patients, indicating that activation of the lectin pathway in AMI patients was much stronger relative to the significantly inhibited PAF patients.
Three pathways share the same late components, including C5, C6, C7, C8 and C9. They together compose the final MAC, which can lyse membrane of bacteria and infected cells, and release inflammatory mediators. It was reported that MAC was associated with unstable coronary diseases (15). When compared with the controls, the significantly elevated mRNA expression of C5 gene, which encodes C5 molecule, may lead to increased formation of MAC in AMI patients, while there was no significant change of mRNA expression of C6, C7, C8 and C9 genes, indicating that the cascade reaction of MAC formation is hard to be continued. In PAF patients, mRNA expression of C5, C6, C8B and C9 genes was significantly downregulated when compared with the controls, indicating there might be a MAC synthesis obstruction in PAF patients. Another researcher also found the reduced MAC level in PAF patients (16). When compared with the PAF patients, the significantly enhanced mRNA expression of C5, C6, C8B and C9 genes in AMI patients indicated that the cascade reaction of MAC formation was stronger in AMI patients than that in PAF patients.
Among the seven genes related to complement receptors, CR1 gene encodes complement receptor 1, which combine with its ligand C3b with a high affinity to promote phagocytosis, clearance of immune complexes, and activation of B lymphocytes (17). Complement receptor 2, encoded by CR2 gene, mainly expresses on the surface of mature B lymphocytes, and combines with opsonized C3b and IgM to enhance immune responses mediated by B lymphocytes (18). ITGAM and ITGAX genes encode complement receptors 3 and 4 respectively, which can combine with ITGB2 into integrin β2. As a receptor of iC3b, it is involved in multiple processes of phagocytosis (19,20). What encoded by C5AR1 gene is C5aR1, which combines with its ligand C5a to help C5a act as an anaphylatoxin and chemokine (21). When compared with the control group, mRNA expression of CR1, ITGAM, ITGAX, ITGB2 and C5AR1 genes was significantly upregulated in AMI patients, indicating that phagocytosis, clearance of immune complexes, and inflammatory responses were enhanced, while significantly downregulated mRNA expression of CR2 gene suggested significantly weakened immune responses mediated by B lymphocytes in AMI patients. An in vivo study also showed that complement receptor 1 inhibitor could reduce the myocardial infarction size (22). But in PAF patients, the significantly decreased mRNA expression of C5AR1 and CR2 genes indicated that the functions of C5a as an anaphylatoxin and chemokine, and immune responses mediated by B lymphocytes were significantly decreased. As for the comparison between AMI and PAF patients, mRNA expression of CR1, ITGAM, ITGAX, ITGB2 and C5AR1 genes was significantly upregulated in AMI patients, indicating that phagocytosis mediated by complements, clearance of immune complexes, and inflammatory responses were significantly stronger than those in PAF patients. However, immune responses mediated by B lymphocytes in both AMI and PAF patients were weaker than that in the controls.
In this study, 10 genes related to regulatory factors affecting the complement system were detected, including C4BPA, C4BPB, CFI, CFH, CFHR1, CD46, CD55, VTN, CLU and CD59. Among them, CFI and CFH encode factor I and factor H respectively, which can combine with C3b to inactivate it into iC3b. And CFHR1 encodes factor H related protein 1. In this study, when compared with the controls, mRNA expression of CFI, CFH and CFHR1 genes did not change significantly, meanwhile mRNA expression of C3 gene did not change significantly either in AMI patients, suggesting no significant negative regulative effect of factor I and factor H on C3b in AMI patients. Other studies did not find any association between factor H and AMI either (23,24). However, mRNA expression of CFI and CFHR1 genes was significantly downregulated, but mRNA expression of C3 gene did not change significantly in PAF patients when compared with the controls, indicating that there was no need for factor I and factor H to inhibit C3b in PAF patients. Thus, it causes the downregulation of mRNA expression of relative genes. As for the comparison between AMI and PAF patients, the significantly upregulated mRNA expression of CFI and CFHR1 genes in AMI patients indicates a stronger inhibitory effect of factor I and factor H on C3b in AMI patients than that in PAF patients.
The protein encoded by CD46 gene assists factor I to degrade C3b and C4b. The protein encoded by CD55 gene can inhibit activities of C3 convertase and C5 convertase. The protein encoded by CD59 gene can block the combination of C7 and C8 with C5b to inhibit the generation of MAC. Protein S encoded by VTN gene can interfere with the combination of C5b67 to cell membrane, leading to a loss of membrane binding activities of C5b-7 and thus obstacles in the formation of MAC. Besides, clusterin encoded by CLU gene can inhibit the assembly of MAC, and promote the dissociation of MAC from cell membrane. When compared with the control group, mRNA expression of CD46, CD55 and CD59 genes was significantly upregulated in AMI patients, indicating that the degradation of C3b and C4b, the main components of C3 convertase and C5 convertase, increased significantly, and thus the activities of C3 convertase and C5 convertase were significantly inhibited. It can be speculated from the simultaneously activated and inhibited complement system that the complement system is not activated actually, but in a disturbed status in AMI patients. However, in PAF patients, mRNA expression of CD46, CD55, CD59 and VTN genes was significantly downregulated when compared with the controls, indicating that the previously inhibitory functions were weakened. Thus, it can be concluded that the inhibitory factors did not function enough due to the significantly decreased complement system function in PAF patients. As for the comparison between AMI and PAF patients, mRNA expression of CD46, CD55, CD59, VTN and CLU genes was significantly upregulated in AMI patients, suggesting that the degradation of C3b and C4b, inhibition on activities of C3 convertase and C5 convertase, and interference with MAC combination were stronger in AMI patients than those in PAF patients.
Taken together, the mRNA expression of related genes in the complement system is under a relatively stable state in SAP patients. Taken SAP patients as the controls, results show that the complement system is in a high-intensive disturbance with simultaneous activation and inhibition in AMI patients, indicating that the cascade response of complement system is disturbed, and then the MAC cannot form finally. The mRNA expression of related genes in the complement system is under a status of downregulation in PAF patients, indicating that the functions of cascade response in the complement system significantly decreased in PAF patients, leading to significantly decreased MAC functions. In conclusion, results of this study indicate that the functions of complement system are in an integrally sthenic imbalance in AMI patients, while the functions of complement system decrease entirely in PAF patients. Thus, several steps, especially late components regarding MAC, in the complement cascade are candidates for therapeutic intervention in AMI. Since traditional Chinese medicine has a role in improving complement system and immune function, it might be a new choice of AMI and PAF treatment.
Acknowledgments
Funding: This study was supported by Shanghai Sailing Program (19YF1444100), the Fundamental Research Funds for the Central Universities (No. 22120180613) and the National Natural Science Foundation of China (No. 81700316).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm.2020.04.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study has been approved by the Ethics Committee of Tongji Hospital (No. KYSB-2014-054), and informed consent form was also obtained. This study has been performed in accordance with the principles of Declaration of Helsinki.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tan Y, Sheng Z, Zhou P, et al. Plasma Trimethylamine N-Oxide as a Novel Biomarker for Plaque Rupture in Patients With ST-Segment-Elevation Myocardial Infarction. Circ Cardiovasc Interv 2019;12:e007281. [Crossref] [PubMed]
- Torzewski M, Bhakdi S. Complement and atherosclerosis-united to the point of no return? Clin Biochem 2013;46:20-5. [Crossref] [PubMed]
- Badjatiya A, Rao SV. Advances in Antiplatelet and Anticoagulant Therapies for NSTE-ACS. Curr Cardiol Rep 2019;21:3. [Crossref] [PubMed]
- Lappegard KT, Hovland A, Pop GA, et al. Atrial fibrillation: inflammation in disguise? Scand J Immunol 2013;78:112-9. [Crossref] [PubMed]
- Duan Q, Gong Z, Song H, et al. Symptomatic venous thromboembolism is a disease related to infection and immune dysfunction. Int J Med Sci 2012;9:453-61. [Crossref] [PubMed]
- Yan W, Wang L, Jiang J, et al. Differential expression of T cell-related genes in AMI and SA stages of coronary artery disease. Int J Clin Exp Med 2015;8:10875-84. [PubMed]
- Halas YA, Rahal E, Abdelnoor AM, et al. Serum C-reactive protein and complement proteins in patients with acute myocardial infarction. Immunopharmacol Immunotoxicol 2005;27:405-16. [Crossref] [PubMed]
- Yan W, Song Y, Zhou L, et al. Immune Cell Repertoire and Their Mediators in Patients with Acute Myocardial Infarction or Stable Angina Pectoris. Int J Med Sci 2017;14:181-90. [Crossref] [PubMed]
- Cubedo J, Padro T, Badimon L. Coordinated proteomic signature changes in immune response and complement proteins in acute myocardial infarction: the implication of serum amyloid P-component. Int J Cardiol 2013;168:5196-204. [Crossref] [PubMed]
- Kishida K, Nakagawa Y, Kobayashi H, et al. High serum C1q-binding adiponectin levels in male patients with acute coronary syndrome. Cardiovasc Diabetol 2014;13:9. [Crossref] [PubMed]
- Szeplaki G, Varga L, Fust G, et al. Role of complement in the pathomechanism of atherosclerotic vascular diseases. Mol Immunol 2009;46:2784-93. [Crossref] [PubMed]
- Nilsson B, Hamad OA, Ahlstrom H, et al. C3 and C4 are strongly related to adipose tissue variables and cardiovascular risk factors. Eur J Clin Invest 2014;44:587-96. [Crossref] [PubMed]
- Distelmaier K, Adlbrecht C, Jakowitsch J, et al. Local complement activation triggers neutrophil recruitment to the site of thrombus formation in acute myocardial infarction. Thromb Haemost 2009;102:564-72. [Crossref] [PubMed]
- Vengen IT, Madsen HO, Garred P, et al. Mannose-binding lectin deficiency is associated with myocardial infarction: the HUNT2 study in Norway. PLoS One 2012;7:e42113. [Crossref] [PubMed]
- Hoffmeister HM, Ehlers R, Buttcher E, et al. Comparison of C-reactive protein and terminal complement complex in patients with unstable angina pectoris versus stable angina pectoris. Am J Cardiol 2002;89:909-12. [Crossref] [PubMed]
- Rubens FD, Nathan H, Labow R, et al. Effects of methylprednisolone and a biocompatible copolymer circuit on blood activation during cardiopulmonary bypass. Ann Thorac Surg 2005;79:655-65. [Crossref] [PubMed]
- Crehan H, Holton P, Wray S, et al. Complement receptor 1 (CR1) and Alzheimer's disease. Immunobiology 2012;217:244-50. [Crossref] [PubMed]
- Giussani S, Pietrocola G, Donnarumma D, et al. The Streptococcus agalactiae complement interfering protein combines multiple complement-inhibitory mechanisms by interacting with both C4 and C3 ligands. FASEB J 2019;33:4448-57. [Crossref] [PubMed]
- Chaves LD, Bao L, Wang Y, et al. Loss of CD11b exacerbates murine complement-mediated tubulointerstitial nephritis. PLoS One 2014;9:e92051. [Crossref] [PubMed]
- Tan SM. The leucocyte beta2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci Rep 2012;32:241-69. [Crossref] [PubMed]
- Moreno-Fernandez ME, Aliberti J, Groeneweg S, et al. A novel role for the receptor of the complement cleavage fragment C5a, C5aR1, in CCR5-mediated entry of HIV into macrophages. AIDS Res Hum Retroviruses 2016;32:399-408. [Crossref] [PubMed]
- Weisman HF, Bartow T, Leppo MK, et al. Soluble human complement receptor type 1: in vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science 1990;249:146-51. [Crossref] [PubMed]
- Sofat R, Casas JP, Kumari M, et al. Genetic variation in complement factor H and risk of coronary heart disease: eight new studies and a meta-analysis of around 48,000 individuals. Atherosclerosis 2010;213:184-90. [Crossref] [PubMed]
- Stark K, Neureuther K, Sedlacek K, et al. The common Y402H variant in complement factor H gene is not associated with susceptibility to myocardial infarction and its related risk factors. Clin Sci (Lond) 2007;113:213-8. [Crossref] [PubMed]


