Genetic biomarkers associated with response to palliative radiotherapy in patients with painful bone metastases
Introduction
Cancer is one of the most prevalent causes of morbidity in Canada, with an estimated 202,400 new cases every year (1). Many cancer patients present with painful bone metastases, which contribute to a reduced quality of life (QOL) and a reduced ability to perform activities of daily living (ADL). When left unmanaged, bone metastases may lead to complications known as skeletal-related events (SREs), such as fractures and spinal cord compression that cause severe pain and functional impairment (2). As cancer treatments have improved the overall survival of cancer patients, patients are dealing with bone metastases on a long term basis (3). For example, 95% of breast and prostate cancer patients survive for at least 5 years or more after diagnosis (4). Therefore, effective pain management of bone metastases in improving QOL and restoring functional independence is an increasingly relevant aspect in cancer palliation.
Palliative radiotherapy (RT) is often used in conjunction with analgesics, hormone treatments, and bone-modifying agents to manage pain secondary to bone metastases (2). However, patients vary in their responses to palliative RT in terms of pain reduction. As defined by the International Bone Metastases Consensus Working Party, responses to palliative RT while considering analgesic dose is categorized into complete response [(CR) complete reduction in pain], partial response [(PR) partial reduction in pain], or pain progression [(PP) increase in pain] (5). Overall, 58–59% of patients have at least a partial pain reduction (CR or PR) (6). Therefore, almost half of patients would not respond to palliative RT and would have either consistent or increased pain levels.
Development of large scale genomic and sequencing technologies have enabled numerous studies to identify and validate genetic biomarkers, which are germline genetic variations in the population that are associated with a biological outcome. Single-nucleotide variants (SNVs) comprise the majority of genetic variation among individuals, and are differences at a single location in DNA (7). While several studies exist on genomic markers predictive of curative radiation treatment or radiation toxicity, genetic biomarkers have not been investigated in a palliative setting (8-11). Therefore, we aimed to identify genetic polymorphisms associated with palliative RT response in patients. These findings may be significant in enabling effective pain management strategies, as well as providing insight into the mechanism of the pain response to RT.
Methods
Patient population
Informed, written consent was obtained for cancer patients across 23 Canadian cancer centres receiving palliative RT of a single 8 Gy dose for painful bone metastases were enrolled in the randomized, double-blind placebo-controlled trial NCIC Clinical Trials Group (NCIC CTG) Symptom Control 23 (SC.23) study (12). This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094).
Data collection
Patients were asked to fill out both the brief pain inventory (BPI) in which they reported their worst pain scores on a scale of 0–10 as well as their opioid analgesic intake on day one of RT, every day for 10 days post-RT, and at day 42 post-RT. Change in pain response from day 1 until day 42 were used to classify RT response based on definitions from the International Bone Metastases Consensus Working Party (5). RT responders consisted of patients who had CR or PR, while poor responders consisted of patients who were non-responders (NR), had PP, or stable pain (SD). Response to RT at week 6 evaluation, CR/PR as responders coded as 1, others coded as 0.
Genomic analysis
Saliva samples were obtained from patients at day of RT. The samples underwent next-generation sequencing using the Illumina TruSightTM One Panel to identify SNVs in 4,813 genes with known disease-causing variants. Raw data from Illumina’s MiSeq platform hg19 was mapped to a reference genome using BWA (13). Base quality score recalibration, indel realignment, duplicate removal, and variant calling using GATK (14) were performed in accordance with the principles outlined in GATK Best Practices (15). Variants with functional and clinical information were annotated using ANNOVAR to assist in subsequent variant filtering and analysis (16).
Variant selection and statistical analysis
Variants were selected from genes identified to be part of inflammatory, immune response, radiation response, or DNA damage. Associations between variants and response to RT were tested for statistical significance using the Cochran-Armitage trend test to produce a univariate model. Significant variants (P<0.005) underwent penalized variable selection to identify a multi-SNVs model predictive of radiation response. The SAS procedure hpgenselect was used with the LASSO method of variable selection using the minimum Bayesian Information Criterion. Each SNVs was coded as 0 if patients had a genotype of AA, 1 for AB and 2 for BB. Patients’ prognostic scores for response to RT at 6 weeks were derived from the sum of the estimate of effect in the hpgenselect model of each of SNVs in the multivariable model, multiplied by the corresponding SNV value (0, 1 or 2). The prognostic score of response to RT at 6-weeks was used to divide patients into three groups: low (<1/3 quantiles), coded as 0 vs. middle (≥1/3 quantiles but <2/3 quantiles), coded as 1 vs. high (≥2/3 quantiles), coded as 2. In multivariable analysis, a logistic regression model was produced with response status as the dependent outcome, and the risk groups model adjusted for gender and primary cancer site as the independent factor.
Pathway analysis was conducted for significant variants and their associated genes to identify genes in commonly reoccurring biological pathways implicated in radiation response. A literature search of significant variants was also conducted and independently analysed for reproducibility of significance.
Results
Baseline demographic and clinical characteristics of the patient population in this study are shown in Table 1. The median age of patients was 72, with the inter-quartile range of 59 to 78 years old. Females represented 41.1% of patients. Prostate was the most common site of primary cancer (31.8%), followed by breast (24.3%) and lung (22.4%). The most common Karnofsky performance status was between 70–80, and the most common worst pain score at baseline was between 7–10. The most common location of radiation treatment was to the pelvis, hips, or lower limbs (40.2%), followed by the ribs, clavicle or sternum (24.3%), then the lumbo-sacral spine (21.5%). Out of 79 patients included in this study, 36 responded to palliative RT (33.6%), and 71 did not (66.4%).
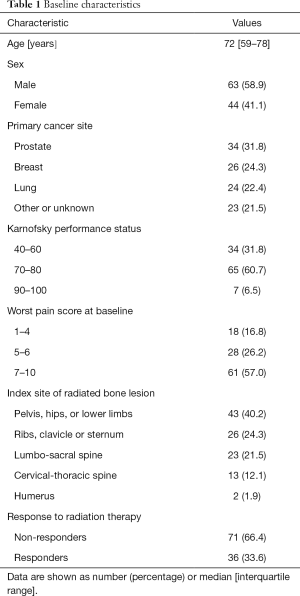
Full table
Multivariable model
Sequencing of 4,813 genes found 41 variants significantly associated with palliative radiation response in univariate analysis (Table S1). The multivariable model selected 14 SNVs (Table 2). A high prognostic score corresponded with a higher chance of response to RT. Univariate analysis of the risk group by response status using the Chi-squared tests showed that 89% of patients in high prognostic group responded to RT (P<0.0001, Table 3).
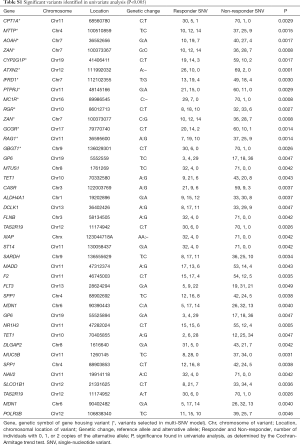
Full table
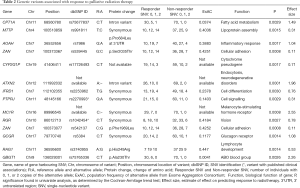
Full table
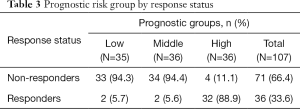
Full table
A deletion variant at position 89986545 on chromosome 16 of the gene MC1R had the largest effect size (2.55). MC1R produced a melanocyte-stimulating hormone receptor. The variant with the highest statistical significance was from an intronic variant of a base deletion at position 111992032 of chromosome 12, corresponding to the gene ataxin (ATXN2, P=0.0001). Our model identified two variants that belonged to a single gene, the cell adhesion gene ZAN. These were the rs539445 variant, which produces an amino acid change at position 2,035 from serine to threonine, and the rs542137 variant, which produces an amino acid change at position 1,969 from phenylalanine to leucine.
Two variants have published associations. The rs2270993 G973A synonymous variant of the cell signalling gene PTPJR was found by Aya-Bonilla et al. to be associated with susceptibility to non-Hodgkin’s lymphoma (17). PTPJC produces a tyrosine phosphatase receptor involved in signal transduction of MAPK signalling of cell growth, proliferation, and angiogenesis, and is putative tumour-suppressor gene. The rs1042454 C:T synonymous variant of the retinal G-protein coupled receptor gene RGR was investigated in two studies. Singh et al. found that it is associated with autosomal recessive retinitis pigmentosa (18). Grupe et al. investigated the association of chromosome 10 variants and late-onset Alzheimer disease (19). However, the RGR variant rs1042454 was not found to be significant.
Discussion
Radiogenomic studies have established that SNV from genes in the pathways of oxidative stress, inflammation, DNA damage signalling and repair, and cell cycle control are involved in determining an individual’s sensitivity to radiation (20). In radiation therapy, ionizing radiation causes double-strand breaks in the DNA, leading to downstream repair mechanisms and signals to the cell to stop cell cycle progression or to undergo apoptosis. Since cancer cells have greater genomic instability, radiation can cause DNA damage to an irreparable degree in these cells, leading to tumour shrinkage. In the palliative setting, radiation can reduce pain through shrinking tumours or metastases that compress nerves or activate pain transducing afferent neurons (21). Therefore, response to palliative RT also involves cytotoxic response to DNA damage in tumour cells.
Our study identified a variant, rs3740955 from the from the RAG1 gene corresponding to a change from histidine to arginine at position 249, that is implicated in response to palliative RT. RAG1 is part of the machinery that produces DNA breaks that facilitates VDJ recombination critical in lymphocyte development (22). A study by Gee et al. on patients with breast cancer who had adjuvant radiation therapy after breast-conserving surgery identified markers associated with radiation response as measured by local recurrence and survival (23). The authors found that low expression of RAG1 was significantly associated with local recurrence in multivariate analysis. Genetic variation of RAG1 may influence response to radiation, and confer differences in susceptibility of tumour cells to radiation damage, therefore producing differences in the ability of radiation to reduce pain through causing tumour shrinkage.
Bone metastases cause bone pain through a combination of nociceptor stimulation by growth factors and proinflammatory molecules produced from the tumour environment, and nerve injury from the growing tumour (24). These may be identified as changes in cellular signalling cascades. Our study identified several variants of intracellular and intercellular signalling that may be implicated in the pain response to radiation therapy. This includes the rs2270993 variant in PTPRJ, a tumour suppressor gene with antiproliferative functions through inhibiting cell growth, migration, and vascularization (25). PTPRJ has also been implicated in several cancer types, including colorectal cancer, non-Hodgkin’s lymphoma, and esophageal squamous cell carcinoma (17,26,27). Another variant that is part of a signalling gene is the deletion at position 89986545 of MC1R. This gene is involved in the development of melanocytes and in sensitivity to solar UV radiation damage (28). Epidemiological studies have also found an association of MCR1 variants with risk of skin cancer.
There are currently several options in managing cancer pain, including radiation, non-steroidal anti-inflammatory drugs, opioids, systemic radioisotopes, corticosteroids, antidepressants, and bisphosphonates. The potential ability of genetic biomarkers to identify patients who respond to palliative RT would allow targeted options for end-of-life care. In the palliative setting, predicting whether a patient will respond to RT would not only allow more efficient management strategies, but also reduce the fatigue and psychological burden from unnecessary treatments that do not produce the desired effect. In addition, it would free-up healthcare resources through serving those patients identified as likely candidates to benefit from RT. Therefore, further research should be conducted to validate the present multi-SNV model. The identification of genetic biomarkers that stratify patients into responders and NR of palliative RT is of great clinical utility to patients and to healthcare management.
Acknowledgements
We thank the generous support of Joey and Mary Furfari Cancer Research Fund. We thank Dr. Ralph Meyer for his contributions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094) and written informed consent was obtained from all patients.
References
- Canadian Cancer Statistics publication. Available online: http://www.cancer.ca/en/cancer-information/cancer-101/canadian-cancer-statistics-publication/?region=on
- Cai B, Nickman NA, Gaffney DK. The role of palliative external beam radiation therapy in boney metastases pain management. J Pain Palliat Care Pharmacother 2013;27:28-34. [Crossref] [PubMed]
- Sciubba DM, Goodwin CR, Yurter A, et al. A systematic review of clinical outcomes and prognostic factors for patients undergoing surgery for spinal metastases secondary to breast cancer. Global Spine J 2016;6:482-96. [Crossref] [PubMed]
- Leading Causes of Death, Total Population, by Age Group and Sex, Canada, Annual. Statistics Canada. Available online: http://www.cancer.ca/en/cancer-information/cancer-101/cancer-statistics-at-a-glance/?region=on
- Chow E, Wu JS, Hoskin P, et al. International consensus on palliative radiotherapy endpoints for future clinical trials in bone metastases. Radiother Oncol 2002;64:275-80. [Crossref] [PubMed]
- Chow E, Harris K, Fan G, et al. Palliative radiotherapy trials for bone metastases: a systematic review. J Clin Oncol 2007;25:1423-36. [Crossref] [PubMed]
- Lupski JR. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ Mol Mutagen 2015;56:419-36. [Crossref] [PubMed]
- Ghazali N, Shaw RJ, Rogers SN, et al. Genomic determinants of normal tissue toxicity after radiotherapy for head and neck malignancy: a systematic review. Oral Oncol 2012;48:1090-100. [Crossref] [PubMed]
- Kelsey CR, Jackson IL, Langdon S, et al. Analysis of single nucleotide polymorphisms and radiation sensitivity of the lung assessed with an objective radiologic endpoin. Clin Lung Cancer 2013;14:267-74. [Crossref] [PubMed]
- Kerns SL, Kundu S, Oh JH, et al. The Prediction of radiotherapy toxicity using single nucleotide polymorphism-based models: a step toward prevention. Semin Radiat Oncol 2015;25:281-91. [Crossref] [PubMed]
- Herskind C, Talbot CJ, Kerns SL, et al. Radiogenomics: a systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett 2016;382:95-109. [Crossref] [PubMed]
- Chow E, Meyer RM, Ding K, et al. Dexamethasone in the prophylaxis of radiation-induced pain flare after palliative radiotherapy for bone metastases: a double-blind, randomised placebo-controlled, phase 3 trial. Lancet Oncol 2015;16:1463-72. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Aya-Bonilla C, Green MR, Camilleri E, et al. High-resolution loss of heterozygosity screening implicates PTPRJ as a potential tumor suppressor gene that affects susceptibility to non-hodgkin’s lymphoma. Genes Chromosomes Cancer 2013;52:467-79. [Crossref] [PubMed]
- Singh HP, Jalali S, Narayanan R, et al. Genetic analysis of indian families with autosomal recessive retinitis pigmentosa by homozygosity screening. Invest Ophthalmol Vis Sci 2009;50:4065-71. [Crossref] [PubMed]
- Grupe A, Li Y, Rowland C, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet 2006;78:78-88. [Crossref] [PubMed]
- Niu N, Qin Y, Fridley B, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 2010;20:1482-92. [Crossref] [PubMed]
- Hayashi S, Tanaka H, Hoshi H. Palliative external-beam radiotherapy for bone metastases from hepatocellular carcinoma. World J Hepatol 2014;6:923-9. [Crossref] [PubMed]
- Notarangelo LD, Kim MS, Walter JE, et al. Human RAG mutations: biochemistry and clinical implications. Nat Rev Immunol 2016;16:234-46. [Crossref] [PubMed]
- Gee HE, Buffa FM, Harris AL, et al. MicroRNA-related DNA repair/cell-cycle genes independently associated with relapse after radiation therapy for early breast cancer. Int J Radiat Oncol Biol Phys 2015;93:1104-14. [Crossref] [PubMed]
- Mantyh PW, Clohisy DR, Koltzenburg M. Molecular mechanisms of cancer pain. Nat Rev Cancer 2002;2:201-9. [Crossref] [PubMed]
- Toland AE, Rozek LS, Presswala S, et al. PTPRJ haplotypes and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 2008;17:2782-5. [Crossref] [PubMed]
- Qiao D, Li M, Pu J, et al. Loss of protein tyrosine phosphatase receptor j expression predicts an aggressive clinical course in patients with esophageal squamous cell carcinoma. Pathol Oncol Res 2016;22:541-7. [Crossref] [PubMed]
- Zhang XF, Tu R, Li K, et al. Tumor Suppressor PTPRJ Is a Target of miR-155 in Colorectal Cancer. J Cell Biochem 2017;118:3391-400. [PubMed]
- Herraiz C, Garcia-Borron JC, Jiménez-Cervantes C, et al. MC1R signaling. Intracellular partners and pathophysiological implications. Biochim Biophys Acta 2017;1863:2448-61. [Crossref] [PubMed]


