
The gut microbiome can be used to predict the gastrointestinal response and efficacy of lung cancer patients undergoing chemotherapy
Introduction
Lung cancer has the highest incidence and mortality rate of any tumor in the world. Despite the continuous advances in lung cancer treatment, patient outcomes are still unsatisfactory, and the 5-year survival rate is a meager 10–20% (1). Globally, approximately three-quarters of lung cancer cases are related to smoking, with the remainder caused by occupational workplace exposure, radon exposure, and air pollution (2).
The treatment approach for lung cancer is mainly determined by the molecular subtype and clinical stage. According to the pathological classification, lung cancer can be divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), with NSCLC accounting for 80–85% of all lung cancer cases. NSCLC can be further divided into histological subtypes, of which adenocarcinoma and squamous-cell carcinoma (SCC) comprise 70% and 20% of cases, respectively (3). For patients with early lung cancer, surgical resection is the standard treatment. For advanced NSCLC patients, there are also many options. In recent years, the rapid development of targeted therapy and immunotherapy has improved treatment strategies for patients with advanced NSCLC. Drugs targeting common driver gene mutations in NSCLC, such as EGFR (epidermal growth factor receptor) mutations, ALK (Anaplastic lymphoma kinase) rearrangements, and KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) mutations, have been developed and have achieved clinical efficacy. Moreover, in the past 2 years, developments in immunotherapy have achieved excellent results in clinical practice. The detection of the expression level of programmed death ligand-1 can be used to guide the treatment of immunotherapy. Despite these advances, platinum-based two-drug combination chemotherapy regimens remain the standard treatment for patients with advanced NSCLC. However, attention should be paid to the adverse reactions associated with this treatment, especially myelosuppression and gastrointestinal mucositis. The adverse reactions often lead to delayed and reduced medication.
The microbiome is a microbial community living in a specific environment, including bacteria, archaea, viruses, and some single-cell eukaryotes (4). There are an estimated 1×1014 colonized bacteria in human microbes, and each individual person has more than 160 bacteria and millions of genes (5-8). Among the bacteria in this huge population, those that live and settle in the gastrointestinal tract are referred to as intrinsic bacteria, and those that only temporarily pass through the gastrointestinal tract are known as heterologous bacteria. According to their concentration, the intrinsic bacteria in the gastrointestinal tract can be classified as dominant bacteria (>107 CFU/g) or inferior bacteria (<107 CFU/g) (9,10). In the small intestine, the density of human gut microbiome is relatively low; however, it gradually increases from the duodenum to the ileocecal, and reaches its highest concentration in the colon (4,11,12). Regional differences in gut microbial populations are caused by functional heterogeneity in various segments of the gastrointestinal tract (13).
Humans have two genomes, one that is inherited from the parents, and the other being the microbiome. The most important difference between these two genomes is that the genetic genome remains almost stable during the life cycle. The microbiome is a complete collection of all the genomic elements of a specific microorganism. The gut microbiome can hold as many as 1,000 bacterial species and encodes approximately 5 million genes, which can perform numerous functions essential to the host’s physiology and survival (4). The gut microbiome is essential for human physiology, immune system development, digestion, and detoxification reactions. The proteins encoded by certain microorganisms found in the gut are essential to the health of the host, for example, the microbial fermentation of complex non-digestible dietary carbohydrates and host-derived glycans (14). However, the microbiome is extremely dynamic and may be influenced by many factors, including age (15), diet (16), hormone cycle (17), physical health, and medications (18). Gut microbiome play a vital role in innate and acquired immune responses throughout life, regulating inflammation, infection, and the balance between food and food antigen tolerance. The colonization and composition of the gut microbiome can influence the immune system and thus can affect various diseases, including asthma, allergies, and inflammatory bowel disease (19). The role played by the gut microbiome in cancer treatment is becoming increasingly clearer (20-23), and evidence suggests that the regulation of gut microbes may affect the response to multiple types of cancer treatment (24-27). The microbiome has received considerable attention owing to its impact on many human diseases, including cancer.
A significant proportion of the global cancer burden is attributable to microbial pathogens, such as human papillomavirus and H. pylori (28,29). An expert group believes that despite the mechanical and supporting evidence obtained from animal and human studies, there is currently no direct evidence that the human symbiotic microbiome is a key determinant of cancer pathogenesis (30). Deep metatranscriptome sequencing analysis of 65 colorectal carcinoma patients showed that in addition to Fusobacterium, Leptotrichia and Campylobacter spp. are co-enriched in colorectal carcinoma (31). Also, to verify the correlation between changes in gut microbiome and the side effects of chemotherapy in cancer, eight patients with non-Hodgkin’s lymphoma who underwent a single course of bone marrow transplantation conditioning chemotherapy were included in the study. The results showed that alpha diversity decreased sharply, and the composition of gut microbiome changed significantly during chemotherapy. Furthermore, the proportion of Faecalibacterium dropped drastically in the patient, while Escherichia increased. Changes in intestinal flora in immunocompromised cancer patients may be closely related to the side effects of chemotherapy (32). In a chemotherapy study of colorectal cancer model rats, irinotecan was found to be able to significantly reduce the total amount of intestinal flora and increase Clostridium cecum and Enterobacteriaceae, especially after intensive treatment (33). Cyclophosphamide was found to considerably reduce the secretion of sIgA in the intestine of mice and lead to the imbalance of the ratio of beneficial bacteria and harmful bacteria (34).
Metagenomic analysis can infer the type of microorganisms in the sample from the presence of its sequence characteristics. It has become a sensitive, culture-independent method for detecting new tumor-associated microorganisms in culture dishes (35). This study aimed to explore the correlation between changes in the gut microbiome and the side effects and efficacy of chemotherapy by conducting a metagenomic analysis of fecal samples collected from 60 lung cancer patients who received chemotherapy for the first time and 17 healthy subjects. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/apm-20-2183).
Methods
Patients
Sixty patients who were diagnosed with lung cancer by cytology or histology between July 2018 and November 2019 were enrolled. All of the patients receiving single or dual drug combination chemotherapy for the first time. None of the patients had a second primary malignancy. Seventeen healthy subjects, who were matched to the lung cancer patients by age and sex, were enrolled as the control group. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional ethics committee of The Second Hospital of Dalian Medical University (No. 41, 2016).
Sample collection
Fecal samples were collected from the study subjects in accordance with the principle of voluntariness, and each subject signed an informed consent form for the collection of samples and data. In lung cancer group, fresh fecal samples were collected from all 60 patients (A1–A60) before chemotherapy and from 37 patients (B1–B37) after chemotherapy. A sample was collected from each of the 17 controls (C1–C17). After collection, the samples were frozen and stored in a refrigerator at –80 °C.
Extraction and purification of fecal DNA
Fecal DNA was extracted using the cetyltrimethylammonium ammonium bromide method. For DNA purification, first, the degree of DNA degradation and potential contamination was assessed on a 1% agarose gel. The DNA concentration was then measured using a dsDNA analysis kit with a Qubit® 2.0 Fluorometer (California Life Technologies). Only DNA samples with an OD (Optical Density) value of 1.8–2.0 and the DNA content above 1 µg were used to construct the library.
Construction of DNA library and sequencing
When the library is constructed, each sample with a total amount of DNA of 1 µg was used as input material for DNA sample preparation. The NEBNext® Ultra™Ultrat was used as input mate (Illumina, NEB, USA) was used to generate a sequencing library and to add an index code to the attribute sequence of each sample. Briefly, DNA sample fragments were sonicated and connected to a full-length adapter for Illumina sequencing, before further amplification by polymerase chain reaction (PCR). Finally, PCR products were purified using the AMPure XP system. The size distribution of the library was analyzed with the Agilent 2100 Bioanalyzer and quantified using real-time PCR. After DNA library passing the test, sequencing was performed on the Illumina HiSeq platform and paired-end reads were generated.
Statistical analysis
Raw data obtained by the Illumina HiSeq sequencing platform were preprocessed with Readfq. If contamination was found, raw data was compared with the host database to obtain valid data. SOAPdenovo software (BGI, China) was used for the assembly of preprocessed data for metagenomic analysis. MetaGeneMark was used to predict the open reading frame of each sample, and redundant data were removed with CD-HIT software (Weizhong Li, Philadelphia, USA). The abundance of information in each sample was calculated. The functional genes were compared with the microbial population extracted from the NR (Non-Redundant) protein sequence database, and the filtered species was analyzed using the LCA (Least Common Ancestors) algorithm to determine the species annotation information of the sequence. Based on the results and gene abundance table, the abundance information and gene number table of each sample at each classification level was obtained. For abundance tables, Krona analysis, PCA (Principal Component Analysis), and NMDS (Nonmetric Multidimensional Scaling) dimensionality reduction analyses were performed. Analysis of similarities was used to test for differences between groups, and then differential species were identified with Metastats and linear discriminant analysis effect size (LEfSe).
Results
Patient characteristics
Sixty lung cancer patients who received chemotherapy for the first time were enrolled in this study to explore the differences in gut microbiome before and after chemotherapy, as well as the correlation between gastrointestinal reactions and gut microbiome. The lung cancer patients in this study had an average age of 63.4 years old, and the majority were male (68%, n=41). Thirty-five patients had a history of smoking, and 13 had received surgical treatment previously. Of the 60 patients in the group, 38 had adenocarcinoma, 13 had SCC, and 9 had SCLC. Gastrointestinal reactions were observed in 26 patients after receiving chemotherapy. Baseline characteristics of the patients are summarized in Table 1.
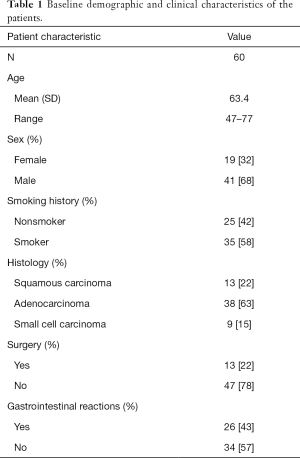
Full table
The gut microbiome of patients with lung cancer differs from that of healthy people
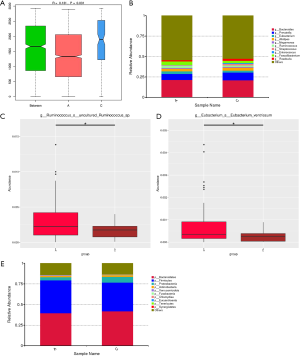
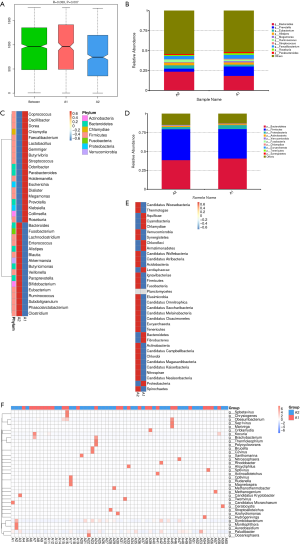
To clarify the difference of gut microbiome between lung cancer patients and healthy individuals, 60 fecal samples (Group A) were collected from the lung cancer patients before chemotherapy and 17 samples (Group C) were collected from the healthy controls. Analysis of similarities based on species abundance was used to assess whether the differences between groups were significantly larger than the variation within groups in order to determine whether the grouping was meaningful. The results (R>0 and P<0.05) indicated that this grouping was meaningful at the genus level (Figure 1A). Based on the relative abundance at the genus level, the top 10 bacterial genera with the largest relative abundance in each sample were identified to be Bacteroides, Prevotella, Eubacterium, Alistipes, Megamonas, Ruminococcus, Streptococcus, Enterococcus, Faecalibacterium, and Roseburia (Figure 1B). Notably, the top 10 bacterial genera with the highest relative abundance were the same between the lung cancer patients and healthy controls. The relative abundance of Eubacterium, Ruminococcus, and Faecalibacterium in lung cancer patients was higher than that in the healthy subjects; however, the lung cancer patients had a lower relative abundance of Prevotella, Streptococcus, Enterococcus, and Roseburia. The abundance difference between Ruminococcus and Eubacterium was found to be the largest through MetaStat Analysis (Figure 1C,D). The top 10 bacterial with the highest relative abundance at the phylum level were also found to be the same between the lung cancer patients and healthy subjects (Figure 1E).
The gut microbiome of lung cancer patients with different pathological types are considerably different
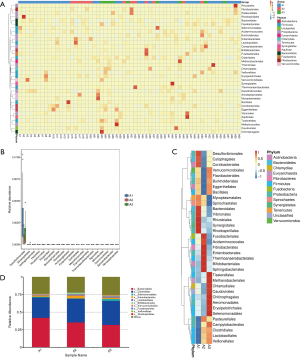
Of the 60 lung cancer patients included in this study, the 38 patients with lung adenocarcinoma were classified as group A1, the 13 patients with SCC were classified as group A2, and the 9 patients with SCLC were classified as group A3. The gut microbial composition was analyzed in these patients at the time of diagnosis, and the relative abundance of each gut microbiota species at the order level was calculated (Figure 2A). As shown in Figure 2B, the composition of the gut microbiome differed significantly between the groups. In the A1 (lung adenocarcinoma) group, there was a relatively high abundance of Desulfovibrionales, Cytophagales, Coriobacteriales, Bacteroidales, Vibrionales, Rhizobiales, Synergistales, and Rhodospirillales. In the A2 (SCC) group, there was a relatively high abundance of Fusobacteriales, Acidaminococcales, Fibrobacteriales, Enterobacteriales, Thermoanaerobacteriales, Bifidobacteriales, and Sphingobacteriales. In the A3 (SCLC) group, there was a relatively high abundance of Tissierellales, Methanobacteriales, Chlamydiales, Caudovirales, Chitinophagales, Aeromonadales, Erysipelotrichales, and Selenomonadales. Next, we analyzed the species with large differences in relative abundance and found that Desulfovibrionales had the largest difference in relative abundance at the order level (Figure 2C). The top 10 species with the largest relative abundance in the three groups were selected. As shown in Figure 2D, the top two species with the highest relative abundance were the same in each group (Bacteroidales and Clostridiales).
Differences in the gut microbiome of lung cancer patients before and after platinum-based chemotherapy
Clustering diagram showing species abundance at the phylum and genus level (Figure 3). In each diagram, the horizontal axis is sample information, and the vertical axis is species annotation information. On the left of the figure is a species clustering tree. The value corresponding to the middle heat map is the Z value obtained by normalizing the relative abundance of each row of species (Figure 3A,B,C). In the relative abundance at the phylum and genus level, the top 10 species with the largest relative abundance in each sample are shown, and “Others” represents other species detected. In the figure, the vertical axis represents the relative proportion of the species annotated to a certain type; the horizontal axis represents the sample name; the species category corresponding to each color block is shown in the legend on the right (Figure 3D,E). Clustering heat map showing species abundance at the genus level (Figure 3F), and box plots (Figure 3G) showing species with significant differences in abundance.
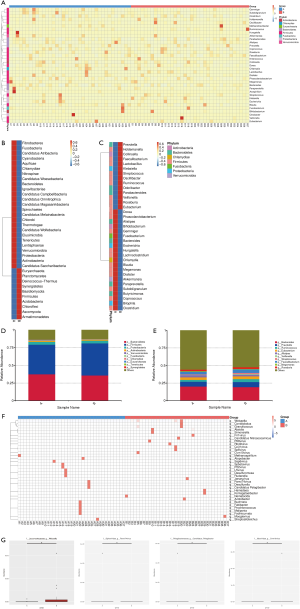
Metagenomics sequencing was applied to assess the composition of intestinal microorganisms in all fecal samples. Fecal samples collected before chemotherapy were classified as A, and those collected after chemotherapy were classified as B. The relative abundance of intestinal bacterial at the levels of phylum, class, order, family, genus, and species was calculated for all lung cancer patients and healthy subjects. At the phylum and genus level, we analyzed the relative abundance of 35 bacteria in feces specimens from lung cancer patients before and after chemotherapy (Figure 3A,B,C). The top 10 bacteria with the largest relative abundance at the phylum and genus levels were identified in all samples and were found to be the same both before and after chemotherapy (Figure 3D,E). However, the relative abundance of each bacterium changed significantly during chemotherapy. At the phylum level, the relative abundance of Firmicutes, Euryarchaeota, and Synergistetes was significantly increased after chemotherapy, while the abundance of Bacteroides, Proteobacteria, Actinobacteria, Verrucomicrobia, Fusobacteria, Chlamydiae, and Tenericutes was significantly decreased (Figure 3D). At the genus level, the relative abundance of Prevotella, Faecalibacterium, Streptococcus, Ruminococcus, Veillonella and Eubacterium was significantly increased after chemotherapy, while the abundance of Bacteroides, Alistipes, Megamonas and Roseburia was significantly decreased (Figure 3E). A significant difference analysis of gut microbial abundance in stool samples before and after chemotherapy was also conducted (Figure 3F). Weissella was the bacterial genus with the significantly difference in abundance before and after chemotherapy at the genus level, followed by Pamx74virus, Candidatus Pelagibacter, and Cvm10virus (Figure 3G). The other 31 bacterial genera with substantial differences in abundance before and after chemotherapy are shown in Figure 3D.
Gut microbiota are associated with gastrointestinal reactions caused by chemotherapy
To examine the relationship between gastrointestinal reactions caused by platinum-based chemotherapy and gut microbiota in lung cancer patients, we classified the 26 patients with gastrointestinal reactions as the A1 group and the 34 patients without gastrointestinal reactions as the A2 group. The rate of gastrointestinal reactions was similar between lung cancer patients with the three pathological types. Of the patients with adenocarcinoma, 42% (16/38) suffered gastrointestinal reactions, compared with 46% (6/13) and 44% (4/9) of SCC and SCLC patients, respectively. Analysis of similarities based on species abundance was used to assess whether the differences between groups were significantly larger than the variation within groups in order to determine whether the grouping was meaningful. The results (R>0 and P<0.05) indicated that this grouping was meaningful at the genus level (Figure 4A). We found that the 10 bacterial genera with the highest relative abundance at baseline were the same whether or not a digestive tract reaction occurred (Figure 4B). Interestingly, our results showed that lung cancer patients with a higher relative abundance of Bacteroides, Eubacterium, Fusobacterium, Lachnoclostridium, Enterococcus, Alistipes, Blautia, Akkermansia, Butyricimonas, Veillonella, Paraprevotella, Bifidobacterium, Ruminococcus, Subdoligranulum, Phascolarctobacterium, or Clostridium at baseline were less likely to have a gastrointestinal reaction (Figure 4B,C). However, patients with a higher relative abundance of Prevotella, Megamonas, Streptococcus, Faecalibacterium, Klebsiella, Roseburia, Lactobacillus, Parabacteroides, Coprococcus, Oscillibacter, Dorea, or Chlamydia, at baseline were more likely to have a gastrointestinal reaction (Figure 4B,C). At the phylum level, patients with a higher relative abundance of Bacteroides, proteobacteria, or Chlamydiae were more likely to experience gastrointestinal reactions. In contrast, a higher relative abundance of Firmicutes, Actinobacteria, Verrucomicrobia, Fusobacteria, Euryarchaeota, Tenericutes, or Synergistetes was associated with a lower risk of gastrointestinal reactions (Figure 4D,E). Also, through a comparative analysis of the gut microbiome of the two groups, 35 bacterial genera with substantial differences in abundance were found (Figure 4F). These results suggested that gastrointestinal reactions caused by chemotherapy were closely related to the gut microbiome.
Gut microbiome can be used to predict the prognosis of lung cancer patients after chemotherapy
Fifty-one of the 60 lung cancer patients in this study were evaluated after 2 cycles of chemotherapy. Forty-one patients with stable disease, partial remission, and complete remission were classified into group A1, and 10 patients with progressive disease were classified into group A2. The relative abundance of gut microbiome before chemotherapy in these 51 patients was analyzed at the genus level (Figure 5A). The results showed that patients with a relatively high abundance of Faecalibacterium, Klebsiella, Coprococcus, Roseburia, Lactobacillus, Streptococcus, Prevotella, Dorea, _or_ Collinsella were more likely to experience disease progression, while patients with a relatively high abundance of Veillonella, Ruminococcus, Paraprevotella, Lachnoclostridium, Akkermansia, or Clostridium were less likely to experience disease progression (Figure 5B). Nevertheless, the two bacterial genera with the highest abundance at the genus level (Bacteroides and Prevotella) were the same in patients with stable disease and those with disease progression (Figure 5C).
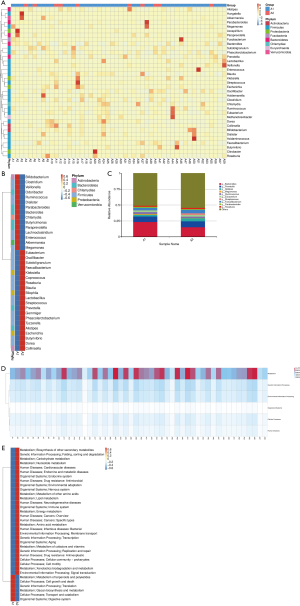
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to associate gene catalogs with higher-level cellular, species, and ecosystem-level system functions. In the KEGG PATHWAY database, biological metabolic pathways are divided into six categories: cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems. We found that the genes associated with the gut microbiome in these lung cancer patients were the highest in metabolic pathways, which indicated that the gut microbiome of lung cancer patients is the most abundant in the metabolic pathway (Figure 5D). Next, we analyzed the gut microbiome of these patients at the second level. Interestingly, the relative abundance of gut microbiome of these two groups of patients was significantly different in different pathways. Patients with a higher relative abundance of enzymes in human diseases pathways were more likely to experience disease progression, while patients with a higher relative abundance of enzymes in the metabolic pathways of terpenoids and polyketides, the cellular processes pathways of cell growth and death, and the genetic information processing pathway of translation were less likely to experience disease progression (Figure 5E). Together, these results show that the gut microbiome can play a role in predicting the prognosis of lung cancer patients after chemotherapy.
Conclusions
A study by Zheng et al. revealed that patients’ microbiome profiles could potentially be used to predict prognosis in early-stage lung cancer (36). Our study uncovered the spectrum of microbiota in lung cancer patients and established the specific gut microbial signature for the potential prediction of the lung cancer. The current study included 60 lung cancer patients and 17 healthy subjects. We compared the abundance and diversity of gut microbiota in the two groups through metagenomic sequencing technology.
The relative abundance of Eubacterium, Ruminococcus, and Faecalibacterium was found to be higher in lung cancer patients than in healthy individuals; however, the opposite relationship was observed in the relative abundance of Prevotella, Streptococcus, Enterococcus, and Roseburia. The abundance of Ruminococcus and Eubacterium in lung cancer was found to be statistically significantly increased at the genus level in lung cancer patients, suggesting that Ruminococcus and Eubacterium may be related to the occurrence and development of lung cancer. Zhuang et al. examined the gut microbiota of 30 lung cancer patients and 30 healthy controls using next-generation sequencing of 16S ribosomal RNA, focusing on microbial diversity and potential biomarkers. They reported no significant decrease in microbial diversity (alpha diversity) in lung cancer patients compared to healthy people, although the composition (beta diversity) differed significantly between the two groups. Healthy individuals were also found to have a higher abundance of the bacterial phylum Actinobacteria and genus Bifidobacterium, while patients with lung cancer showed elevated levels of Enterococcus (37). In the present study, we found that the intestinal microbiota of lung cancer patients with different pathological types also show considerable differences. Zhuang et al.’s findings are very similar to those of the current study, which indicated that gut microbiome was found to be possible biomarkers for lung cancer. These systematic and multi-layered results provide essential information on the role of intestinal microbes in lung cancer, which hold promise for early prevention and targeted intervention for lung cancer patients.
The gut microbiome and its interaction with the host’s innate immune system are believed to play a key role in the development of gastrointestinal reactions caused by chemotherapy; however, comprehensive bioinformatics modeling has not been carried out (38). Furthermore, to verify the correlation between changes in intestinal microbiota and the side effects of chemotherapy, one study enrolled eight patients with non-Hodgkin’s lymphoma who underwent a single course of bone marrow transplantation conditioning chemotherapy. They observed a sharp decrease in alpha diversity during chemotherapy, and the composition of the gut microbiome also changed significantly. Changes in gut microbiome in immunocompromised cancer patients may be closely related to the side effects of chemotherapy (32). Montassier et al.’s experimental results are slightly different from ours (32). In the present study, we found that the relative abundance of Firmicutes, Euryarchaeota, and Synergistetes in lung cancer patients increased significantly after chemotherapy. In contrast, the abundance of Bacteroides, Proteobacteria, Actinobacteria, Verrucomicrobia, Fusobacteria, Chlamydiae, and Tenericutes decreased significantly at the phylum level. At the genus level, the relative abundance of Prevotella, Faecalibacterium, Streptococcus, Ruminococcus, Veillonella, and Eubacterium in lung cancer patients increased significantly after chemotherapy, while an abundance of Bacteroides, Alistipes, Megamonas, and Roseburia showed a considerable decrease. This shows that the gut microbiome of lung cancer patients changes dramatically after treatment with cytotoxic drugs, which may be related to the gastrointestinal reaction caused by chemotherapy. Our results showed that lung cancer patients with a higher relative abundance of Bacteroides, proteobacteria, or Chlamydiae were more likely to have gastrointestinal reactions at the phylum level. In contrast, patients with a higher relative abundance of Firmicutes, Actinobacteria, Verrucomicrobia, Fusobacteria, Euryarchaeota, Tenericutes, or Synergistetes were less likely to have gastrointestinal reactions. Similarly, at the genus level, lung cancer patients with a higher relative abundance of Bacteroides, Eubacterium, Fusobacterium, Lachnoclostridium, Enterococcus, or Alistipes at baseline were less likely to experience gastrointestinal reactions. However, lung cancer patients with a higher relative abundance of Prevotella, Megamonas, Streptococcus, Faecalibacterium, Roseburia, or Parabacteroides, at baseline were more likely to have gastrointestinal reactions.
One of the most common side effects of cancer treatment is diarrhea. The pathogenesis of adverse gastrointestinal reactions can be summarized with the Sonis model. This model includes the following stages: (I) the initial stage, with active oxygen formation; (II) significant injury response to inflammation and apoptosis; (III) signal amplification, in which a cascade reaction promotes inflammation and apoptosis; (IV) ulcer formation, which leads to a discontinuous epithelial barrier, which in turn supports bacterial translocation; (V) treatment, during which cell proliferation stops after chemotherapy or radiotherapy (39). Intestinal microorganisms play a crucial part in maintaining the balance and integrity of the gut. Probiotics to prevent mucositis caused by cancer treatment have been studied in preclinical trials and randomized clinical trials, and have achieved some promising results. Probiotic mixture has been reported to reduce the weight loss of mice before and after irinotecan treatment, and can also prevent moderate and severe diarrhea. These effects are related to a significant increase in crypt proliferation and the suppression of apoptosis in cells of the small and large intestine. The addition of probiotics also prevented irinotecan from inducing an increase in goblet cells in jejunal crypts. Probiotic mixture can also effectively prevent severe diarrhea after irinotecan chemotherapy, so may be used to treat cancer patients (40).
Most importantly, we found in this study that lung cancer patients with a relatively high abundance of Faecalibacterium, Klebsiella, Coprococcus, Roseburia, Lactobacillus, Streptococcus, Prevotella, Dorea, or Collinsella are not only susceptible to gastrointestinal reactions, but are also prone to disease progression after two cycles of chemotherapy. Moreover, the results of the study show that if the relative abundance of Veillonella, Ruminococcus, Paraprevotella, Lachnoclostridium, Akkermansia, Clostridium in the gut microbiome of lung cancer patients is high, then the possibility of gastrointestinal reactions is low, and the risk of disease progression is lower. Therefore, our experimental results suggest that the use of microbial preparations to reduce gastrointestinal mucositis caused by chemotherapy is feasible and could improve the effect of chemotherapy in cancer patients. However, due to the heterogeneity of the human body, some studies have found that the application of microbial preparations increases the risk of infection for patients. Thus, further in-depth exploration of the microbial composition in lung cancer patients is needed to provide theoretical support for individualized treatment in the future.
Acknowledgments
Funding: This work was supported by the Dalian Science and Technology Innovation Fund (2020JJ27SN090).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-2183
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-2183
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-2183). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics committee of The Second Hospital of Dalian Medical University (No. 41, 2016), and written informed consents were obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Malhotra J, Malvezzi M, Negri E, et al. Risk factors for lung cancer worldwide. Eur Respir J 2016;48:889-902. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006;7:688-93. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016;164:337-40. [Crossref] [PubMed]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65. [Crossref] [PubMed]
- Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017;550:61-6. [Crossref] [PubMed]
- Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355-9. [Crossref] [PubMed]
- Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009;9:123. [Crossref] [PubMed]
- Ducluzeau R, Cerf M, Corthier G, et al. Microbial Ecology and Intestinal Infections. Springer Science & Business Media, 2013.
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004;4:478-85. [Crossref] [PubMed]
- Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun 2005;73:3197-209. [Crossref] [PubMed]
- Martinez-Guryn K, Leone V, Chang EB. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019;26:314-24. [Crossref] [PubMed]
- Flint HJ, Scott KP, Duncan SH, et al. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012;3:289-306. [Crossref] [PubMed]
- Trosvik P, de Muinck EJ, Stenseth NC. Biotic interactions and temporal dynamics of the human gastrointestinal microbiota. ISME J 2015;9:533-41. [Crossref] [PubMed]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559-63. [Crossref] [PubMed]
- Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470-80. [Crossref] [PubMed]
- Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013;62:1591-601. [Crossref] [PubMed]
- Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science 2016;352:539-44. [Crossref] [PubMed]
- von Frieling J, Fink C, Hamm J, et al. Grow With the Challenge - Microbial Effects on Epithelial Proliferation, Carcinogenesis, and Cancer Therapy. Front Microbiol 2018;9:2020. [Crossref] [PubMed]
- Heshiki Y, Vazquez-Uribe R, Li J, et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020;8:28. [Crossref] [PubMed]
- Si H, Yang Q, Hu H, Ding C, et al. Colorectal cancer occurrence and treatment based on changes in intestinal flora. Semin Cancer Biol 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Zheng DW, Dong X, Pan P, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng 2019;3:717-28. [Crossref] [PubMed]
- Gopalakrishnan V, Helmink BA, Spencer CN, et al. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018;33:570-80. [Crossref] [PubMed]
- Münch NS, Fang HY, Ingermann J, et al. High-Fat Diet Accelerates Carcinogenesis in a Mouse Model of Barrett’s Esophagus via Interleukin 8 and Alterations to the Gut Microbiome. Gastroenterology 2019;157:492-506.e2. [Crossref] [PubMed]
- Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 2020;17:53-64. [Crossref] [PubMed]
- Picardo SL, Coburn B, Hansen AR. The microbiome and cancer for clinicians. Crit Rev Oncol Hematol 2019;141:1-12. [Crossref] [PubMed]
- Hold GL, Hansen R. Impact of the Gastrointestinal Microbiome in Health and Disease: Co-evolution with the Host Immune System. Curr Top Microbiol Immunol 2019;421:303-18. [Crossref] [PubMed]
- Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120-7. [Crossref] [PubMed]
- Scott AJ, Alexander JL, Merrifield CA, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019;68:1624-32. [Crossref] [PubMed]
- Warren RL, Freeman DJ, Pleasance S, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013;1:16. [Crossref] [PubMed]
- Montassier E, Batard E, Massart S, et al. 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol 2014;67:690-9. [Crossref] [PubMed]
- Lin XB, Dieleman LA, Ketabi A, et al. Irinotecan (CPT-11) chemotherapy alters intestinal microbiota in tumour bearing rats. PLoS One 2012;7:e39764. [Crossref] [PubMed]
- Zuo T, Cao L, Li X, et al. The squid ink polysaccharides protect tight junctions and adherens junctions from chemotherapeutic injury in the small intestinal epithelium of mice. Nutr Cancer 2015;67:364-71. [Crossref] [PubMed]
- Moore RA, Warren RL, Freeman JD, et al. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One 2011;6:e19838. [Crossref] [PubMed]
- Zheng Y, Fang Z, Xue Y, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020;11:1030-42. [Crossref] [PubMed]
- Zhuang H, Cheng L, Wang Y, et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front Cell Infect Microbiol 2019;9:112. [Crossref] [PubMed]
- Secombe KR, Coller JK, Gibson RJ, et al. The bidirectional interaction of the gut microbiome and the innate immune system: Implications for chemotherapy-induced gastrointestinal toxicity. Int J Cancer 2019;144:2365-76. [Crossref] [PubMed]
- Sonis ST. The pathobiology of mucositis. Nat Rev Cancer 2004;4:277-84. [Crossref] [PubMed]
- Bowen JM, Stringer AM, Gibson RJ, et al. VSL#3 probiotic treatment reduces chemotherapy-induced diarrhea and weight loss. Cancer Biol Ther 2007;6:1449-54. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


