
Negative results in nucleic acid test of COVID-19 patients: assessment from the perspective of clinical laboratories
Since December 2019, there had been an outbreak of a novel coronavirus-induced pneumonia across the world. Until February 14, 2020, China had 66,383 confirmed cases already. On February 11, 2020, the World Health Organization (WHO) named the novel coronavirus-induced pneumonia as coronavirus disease 2019 (COVID-19). At the same time, the International Committee on Taxonomy of Viruses named this pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Guidelines issued by the National Health Commission of the People’s Republic of China proposed that one of the diagnostic criteria be positive result of nucleic acid test of SARS-CoV-2 in respiratory or blood samples of patients (1). To respond to the needs of outbreak prevention, rapid diagnosis, and disease monitoring, companies and clinical laboratories have quickly developed nucleic acid test kits for SARS-CoV-2 and put them into clinical application. However, with an increasing number of false-negative results of nucleic acid test and the serious consequences therefore (1), first-line clinicians are starting to believe that nucleic acid test is not reliable, and they have been recommended to use imaging results as a more accurate auxiliary diagnostic tool (1). Researchers believe that the positive rate of nucleic acid test of SARS-CoV-2 is approximately 30–50% (2-4). In this study, we analyzed the causes of false-negative results in nucleic acid test in COVID-19 from the perspective of clinical laboratory and explored measures accordingly.
Possible causes of false-negative results
Pretest processes
Sample collection
Quality of microbial samples affect results strongly. Respiratory tract samples are often used. Upper-respiratory-tract samples include nasopharyngeal and/or oropharyngeal swabs and aspirates, and lower-respiratory-tract samples include phlegm, respiratory aspirates, bronchoalveolar lavage fluid, and lung biopsy samples. According to experience from SARS, viral loads and genome fractions in lower-respiratory-tract samples are high (5), so lower-respiratory-tract samples should be used for testing, followed by nasopharyngeal and/or oropharyngeal swabs. It was reported in some cases that nucleic acid test results of swabs were negative before hospitalization, whereas diagnosis were eventually confirmed by using bronchoalveolar lavage fluid in a few cases (6). However, collection of lower-respiratory-tract samples is very difficult. Due to the specific clinical conditions as well as patient intolerance (for example, the sputum sample should be obtained by deep coughing after washing oral cavity, while many severely ill patients become very weak and fail to provide qualified samples but mostly saliva), nasopharyngeal and oropharyngeal swabs are the mainly used samples for nucleic acid test.
On the other hand, reason for the low positive rate of nucleic acid test in upper-respiratory-tract samples is largely due to unqualified samples. Sampling of oropharyngeal swabs demands expertly operators. Swabs should be scraped from deep down the isthmus faucium around the uvula and palatine tonsils repeatedly (7-9). Some medical personnel, who lack experience and have greater mental stress than usual, fail to collect the samples in the right place with the optimal way, and are unable to obtain perfect epithelial cells. In addition, patients are too sensitive and become uncooperative during sampling. Nasopharyngeal swabs should be acquired by scraping in the deep nasal cavity repeatedly (10). In practice, the positive rate of nucleic acid test using nasopharyngeal swabs is higher than that using oropharyngeal swabs (11,12). Therefore, Diagnosis and treatment of COVID-19 (fifth edition) has changed the oropharyngeal swabs to nasopharyngeal swabs (13). Experts suggest that the combined use of multiple respiratory-tract samples of the patients in testing improve the positive test rate. Besides, patients who have just coughed sputum out or been treated for respiratory secretions may have low viral loads in sampling, leading to false-negative results; swab sampling quality affect the elution of epithelial cells, and dried swabbing after sampling affect nucleic acid extraction later.
Transport of samples
SARS-CoV-2 is RNA virus that could easily be degraded by RNA enzymes exogenous or being released after cell lysis (1). According to the Guidance on the Application of Accreditation Criteria for the Medical Laboratory Quality and Competence in the Field of Molecular Diagnostics (CNAS-CL02-A009) (14) issued by the China National Accreditation Service for Conformity Assessment (CNAS), blood samples for RNA amplification test should be treated with anticoagulation, and plasma should be separated as soon as possible; if anticoagulation is not available, serum should be separated at once. Samples should be tested within 2–4 hours (15), stored at 4 °C for 72 hours at most. Samples and the extracted nucleic acids should be stored below −70 °C if tests cannot be run in time (16). Long storage time and nonstandard temperature affect the test results.
Factors during analysis
Kit
Currently, reverse transcription-real-time fluorescence polymerase chain reaction (RT-PCR) is commonly used in clinical laboratories or centers for disease control and prevention (CDCs). This method has been widely used in routine tests and scientific research experiments. SARS-CoV-2 test kits developed by most companies are based on this method. Under normal circumstances, clinical reagent kits need to be repeatedly verified and evaluated on many samples from development to actual application. However, this outbreak is severe, and time is tight. Many reagent kits are directly applied in clinical laboratories without quality and parameter evaluation. Many experts have observed differences in the positive rate between different kits (1).
First, close attention should be paid to the extraction and preparation of nucleic acid (17) since quality of nucleic acid extracted directly affects RT-PCR results. It can be affected by extracted amount, loading amount, methodology and various processes (18,19). For instance, the loading amount is usually evaluated by the liquid volume instead of the actual amount of RNA, which may lead to the difference of quality of RNA extracted using various methods, affecting amplification results. Expert consensus documents recommend that specimens should be stored in special tubes containing preserving liquid (Guanidine thiocyanate is preferred for ideal inactivation effect and enhanced positive rate) (20) before inactivation at 56 °C for 30 min (21) or 60–65 °C for 20 min (10). Robust evidence has been provided that concentration and purity of nucleic acid extracted by different kits are statistically significant (22), with Ct values of nucleoprotein (N) gene results being statically different. Li mentioned that applied SARS-CoV-2 extraction means at present are mainly direct lysis method, column method and magnetic bead method, among which the last one owned the highest efficiency as well as reduced false negative rate (18).
Three specific regions of SARS-CoV-2 are for RT-PCR: open reading frame (ORF) 1ab, the nucleoprotein (N) gene, and the envelope (E) gene (21). CDCs consider ORF1ab, a confirmation target, presenting the highest specificity; N is an additional confirmation target, while E being a first-line screening target (23). Most kits target ORF1ab as well as N or E gene, also with internal reference sequence monitoring extraction, reverse transcription and quantitative test. Cycling conditions in most Chinese laboratories were as follows (Shengxiang Biotechnology Co., LTD): hold for 30 mins at 50 °C (reverse transcription), hold for 1 min at 95 °C (initial denaturation of cDNA), then 45 cycles of 95 °C for 15 s, 60 °C for 30 s, and 25 °C for 10 s (denaturation, annealing and extension, cooling of equipment). Samples with Ct value (ORF 1ab as well as N gene) ≤40, showing the “S” amplification curve are considered positive. Negative control should be presenting no Ct value or Ct value <40; positive control should be presenting Ct value ≤35. “Suspicious case” is defined as either ORF1ab or N gene is positive, re-sample and recheck with a different kit would be required. Some expert call for attention of “Indefinite Range”, which is defined as Ct value falls within 37–40 (24). If only result of one gene falls within indefinite range, RNA should be re-extracted, amplified and detected with the previous extracted nucleic acid simultaneously. If a single site is positive twice or the two sites are positive, reported; if both results are in the “gray area”, a “negative” is reported. Low viral load extracted, limits, interfering substance may lead to false-negative results (22). RNA viruses have high genetic variability (25). Mismatches between primers, probes, and target sequences caused by gene mutations can lead to suspect or false-negative results. However, the mutation rate of SARS-CoV, which has high homology with SARS-CoV-2, is very low (26). Although there is no relevant report yet, the clinical laboratories should pay close attention to this issue and perform gene sequencing if necessary.
Expert proposed that internal quality control be at least 1 weakly positive control (usually 3 times the test limit) and 3 negative controls (usually 2 from the kit, the other one being a saline) for each batch, randomly placed among clinical specimens. Record Ct value of the weakly positive control in every test. As for external quality assessment (EQA), institutions should be qualified in EQA and biosafety supervision before conducting SARS-CoV-2 experiments for the first time with assessment of provincial clinical testing centers at least once a year (20).
Timing of sampling
The disease has a long incubation period, and most patients were exposed several days to several weeks before the diagnosis. The different viral loads in patients with different clinical types and differences in the amount of detoxification between courses of disease can cause false-negative results (12). The associations between viral load, symptoms, and disease course are valuable information needed for clinical collaboration feedback (1). For one patient, the result of nucleic acid test using oropharyngeal swabs was negative for 2 consecutive days, but that for fecal samples was positive (27). Researchers believe that due to the low viral loads in the mouth/nasopharynx of patients at an early stage or patients with mild symptoms, fecal samples might be more ideal for examination (3,28).
Laboratory capability
CNAS has strict requirements for the Medical Laboratory Quality and Competence in the Field of Molecular Diagnostics (14). Ensuring the capability of molecular diagnostic laboratories is the greatest contribution of laboratory medicine in the control and prevention of outbreak. According to the National Health Committee of the People’s Republic of China, if the laboratory is not qualified, under the premise of biological safety, the samples should be sent to the nearest qualified medical institution for testing, or the samples can be first collected by the county or district CDC, then sent to the provincial CDC or the municipal CDC (16).
Suggestions and measures
Sample selection
Lower-respiratory-tract samples, such as sputum produced by deep coughing, should be considered as long as the biosafety protection is satisfied and patient can tolerate it. For the more often used upper-respiratory-tract samples, such as nasopharyngeal and oropharyngeal swabs, the procedure and method recommended by the Chinese Society of Laboratory Medicine of Chinese Medical Association should be strictly followed (8,21). Samples collected within 3 days of onset are preferred (24). Nasopharyngeal swabs should be collected as follows: a swab is used to measure the distance from the tip of the nose to the earlobes, then mark this distance on the swab with a finger. Insert swab into the nasal cavity perpendicular to the nose (face), and the distance should be at least half of the length from the earlobe to the tip of the nose. The swab stays in the nose for 15–30 s while being gently rotated 3–5 times, and is quickly placed into the sample collection tube containing 2 mL lysis buffer (the same as the lysis buffer in the nucleic acid extraction kit) or containing cell preservation solution with RNase inhibitor. After inserting the swab, the sterile swab rod is broken, and the tube is sealed with film (24,29).
Oropharyngeal swabs are collected as follows: a sterile swab is used to gently swab the back of the pharyngeal area, avoiding touching the tongue. Then place the sample into the collection tube after the rod is broken near the top, and the tube is sealed with film (24,29).
Medical personnel should pay attention to details, such as sampling position, and ensure the best sampling quality. According to fifth edition of the Diagnosis and treatment of COVID-19 (13), multiple nasopharyngeal swabs collected simultaneously are recommended.
Sample transport process
Fast track has been opened for patients with suspected COVID-19 infection in fever clinics, trying to shorten TAT to ensure the quality of samples. The required consumable and biosafety supplies for collection and transport of samples should be carefully verified and processed to avoid the degradation of RNA caused by exogenous RNA enzymes (10). Personnel should be trained about biological protective measures and transportation conditions. For laboratories that are unable to conduct nucleic acid test, necessary conditions should be created for sample transport. These details can greatly improve the accuracy of laboratory diagnosis.
Sample re-examination and laboratory evaluation
Accuracy of the kit is currently the most concerning issue in clinical laboratory and R&D development. The epidemic is out of blue, kits are approved quickly (1), and laboratories lack sufficient data to support the quality assessment. Parallel tests with different reagent kits between and within samples can effectively control false-negative results (22). Samples that still cannot be definitively confirmed should be promptly tested with other kits or by different methods. For highly suspected cases, or the test results are difficult to determine, it is recommended to use more than 2 kits for verification (1).
Some experts point out that nucleic acid test with higher sensitivity (such as by digital PCR) can reduce the false-negative results caused by insufficient sensitivity of RT-PCR (1), which is not popularized owing to the equipments unfortunately. For laboratories that cannot perform nucleic acid test, condition should be improved at best efforts, medical personnel should be trained to share the burden; or samples should be transported properly to qualified laboratories or local CDCs for confirmation. All of the above are summarized in Figure 1.
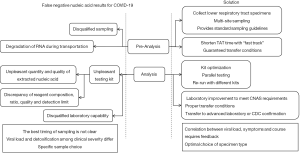
Conclusions
Outbreak of COVID-19 is a huge challenge for both China and the world. Nucleic acid test is an irreplaceable, effective means for the diagnosis of COVID-19. We should standardize sampling, transport materials as soon as possible, ensure the quality of kits, improve the laboratory equipment and laboratory capability, and optimize the test procedures to improve the accuracy of nucleic acid test.
Acknowledgments
Funding: This work was supported by grants s from Jilin Science and Technology Development Program (No. 20170623092TC-09 to JX; No .20190304110YY to JX), The First Hospital Translational Funding for Scientific & Technological Achievements (No. JDYYZH-1902002 to JX).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-568). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mo Q, Qin W, Fu QH, et al. Understanding the Influence Factors in Viral Nucleic Acid Test of 2019 novel Coronavirus (2019-nCoV). Chinese Journal of Laboratory Medicine 2020;(00):E2.
- Huang JT, Ran RX, Lv ZH, et al. Chronological Changes of Viral Shedding in Adult Inpatients with COVID-19 in Wuhan, China. Clin Infect Dis 2020. [Epub ahead of print].
- Yun H, Sun Z, Wu J, et al. Laboratory data analysis of novel coronavirus (COVID-19) screening in 2510 patients. Clin Chim Acta 2020;507:94-7. [Crossref] [PubMed]
- Ma HX, Pan JJ, Li Y, et al. Real-time RT-PCR for the detection of SARS-CoV-2 nucleic acid. Chinese Journal of Microbiology and Immunology 2020.245-6.
- Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018;23:130-7. [Crossref] [PubMed]
- Tan FR, Chou YL, Xu Z. Bronchoalveolar lavage fluid was used to diagnose two cases of 2019-nCoV infection. Chinese Journal of Tuberculosis and Respiratory Diseases 2020;43:337-9. [PubMed]
- Uyeki TM, Bernstein HH, Bradley JS, et al. Clinical Practice Guidelines by the Infectious Diseases Society of America: 2018 Update on Diagnosis, Treatment, Chemoprophylaxis, and Institutional Outbreak Management of Seasonal Influenzaa. Clin Infect Dis 2019;68:895-902. [Crossref] [PubMed]
- Chineses Laboratory Medicine Association. Throat swab specimen collection process of COVID-19. 2020.
- WHO. Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: Interim Guidance. 2020.
- Expert consensus on COVID-19 nucleic acid test. Chinese Medical Journal 2020.968-9.
- Wang X, Tan L, Wang X, et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis 2020;94:107-9. [Crossref] [PubMed]
- Liu P, Cai J, Jia R, et al. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerging Microbes & Infections 2020;9:1254-8. [Crossref] [PubMed]
- National Health Commission of the People's Republic of China. Diagnosis and treatment plan of new coronavirus pneumonia (Trial Version 5, Revised).
- Guidance on the Application of Accreditation Criteria for the Medical Laboratory Quality and Competence in the Field of Molecular Diagnostics. CNAS CL02 A009-2018: 0.
- National Health Commission of the People's Republic of China. Guidelines for laboratory techniques for new coronavirus pneumonia (3rd edition). 2020.
- National Health Commission of the People's Republic of China. Laboratory Biosafety Guidelines of New Coronavirus (2nd edition). 2020.
- Chen PS, He YT, Huang YL, et al. Effect on results of real-time fluorescence quantitative PCR of 2019 new coronavirus using different methods to inactivate oropharyngeal swab specimens. Chinese Journal of Laboratory Medicine 2020;43:364-7.
- Li J, Ye GM, Chen LJ, et al. Analysis of false false-negative results for 2019 novel coronavirus nucleic acid test and related countermeasures. Chinese Journal of Laboratory Medicine 2020;(03):221-2.
- Li RR, Wang B, Hu S, et al. Comparative Evaluation on Different RNA Extraction Methods. Forensic Science and Technology 2018;43:431-5.
- Guan M. Expert consensus on clinical application of new coronavirus nucleic acid and antibody detection. International Journal of Laboratory Medicine.
- Tong YQ, Wang M, Xu WZ, et al. Suggestions on operation for nucleic acid test of new coronavirus in clinical laboratory. 2020.
- Ma W, Zhang WH, Ma YL, et al. Comparative performance of nucleic acid extraction kits for the detection of 2019 novel coronavirus. Journal of Molecular Diagnosis and Therapy 2020;12:552-6.
- Prevention, Centers For Disease Control. Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus. 2020.
- Chinese Laboratory Medicine Association. Expert consensus on nucleic acid test of new coronavirus. Chinese Medical Journal 2020.E3.
- Yang G. Study on genetic drug resistance polymorphism in NS5B region of hepatitis C virus. Guangxi Medical University, 2019.
- Mi W. Progress on the origin and variation of SARS virus. Modern Preventive Medicine 2005;32:749-50.
- Li P, Zhao SL, Chen YF, et al. Clinical implications of 2 cases of coronavirus disease positive for SARS-CoV-2 nucleic acid in fece. International Journal of Laboratory Medicine.
- Han MS, Seong MW, Kim N, et al. Viral RNA Load in Mildly Symptomatic and Asymptomatic Children with COVID-19, Seoul. Emerg Infect Dis 2020;26:2497-99. [Crossref] [PubMed]
- Collection and transfer of clinical microbiology specimens (released version). WS/T 640-2018:24.


