The correlations between the serum expression of miR-206 and the severity and prognosis of sepsis
Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection (1). Sepsis remains a global health problem. Despite recent advances in treatments, including antimicrobials and organ support, the survival rate of patients with sepsis has not been remarkably improved (2). The in-hospital mortality rate of patients with septic shock exceeds 40%. Notably, sepsis pathogenesis is complex and involves multiple mechanisms, including infection, inflammatory response, and immunity. The currently available biomarkers cannot achieve the early diagnosis and tailored treatment of sepsis (especially septic shock). Therefore, biomarkers that can reflect the early occurrence and progression of sepsis and predict its prognosis are urgently needed (3). Serum MiRNAs are a group of non-coding RNA (ncRNA) that functions at the RNA level. As a genetic material, it plays a crucial role in cell-to-cell communication (4). The past few years have witnessed in-depth research on miRNA’s role in various diseases; meanwhile, the potential role of miRNA in sepsis has also been recognized. Some authors have explored the miRNA expression profile on sepsis’s background (5), and its potential role has been investigated. However, the mechanisms via which miRNA participates in the pathogenesis of sepsis deserve further study. In our current study, we used high-throughput sequencing technology to analyze serum miRNA in septic patients and explore the possible correlation between serum miRNA and pathogenic mechanisms of sepsis, with an attempt to find a new biomarker for the early assessment of sepsis severity and prognosis and to pave the way for the tailored treatment of sepsis.
Dysregulated inflammatory response and fatal multiple organ dysfunction are the leading causes of deaths in sepsis. The current routine clinical tests for sepsis include routine blood tests, hypersensitive C-reactive protein (hs-CRP), erythrocyte sedimentation rate (ESR), procalcitonin (PCT), and blood culture (6). Although some of these indicators may reflect the severity of sepsis, they are less effective in the early diagnosis of sepsis (especially septic shock). Even worse, they are not specific and can be easily affected by other factors. An ideal biomarker of sepsis should have a specific correlation with the clinical course of sepsis, and its expression level should have remarkably changed before the levels of the existing biomarkers change.
miRNAs are small, single-stranded non-coding RNAs with a length of 20−22 nucleotides. By binding to the 3'UTR region of the ORF region of mRNA, it acts on specific target genes, mediates post-transcriptional gene silencing, and participates in the pathogenesis of various diseases. More evidence indicates that miRNA plays a vital role in the pathophysiological processes (e.g., cell proliferation, metabolism, and apoptosis and organ development) of many diseases (7). miRNA also plays a vital role in the regulation of immune response in sepsis, as proved in many experiments. Kingsley et al. studied whether miRNA could induce inflammation regulation in sepsis and found the expression levels of miR-182, miR-143, miR-145, miR-146a, miR-150, and miR-155, which are strictly related to immune response, were dysregulated in sepsis patients (8). Thus, miRNA may affect inflammatory response during sepsis. Ma et al. further detected the expression of miR-150 in human umbilical vein endothelial cells (HUVEC) after LPS treatment and found that miR-150 overexpression reduced inflammation and parenchymal cell apoptosis, suggesting MiR-150 may be associated with the severity of sepsis (9). Macrophages are involved in the inflammatory process of sepsis, especially the sepsis-induced acute lung injury (10). Many experiments have demonstrated that miR-9, miR-127, miR-155, and miR-125b can promote macrophage M1 polarization, whereas miR-124, miR-223, miR-34a, miR-132, miR-146a, miR-124b, and miR-125a-5p induce macrophage M2 polarization by targeting various transcription factors to reduce the damage to target organs from sepsis (11-13). An experiment on a miRNA specific for TLR4 found that the miRNA negatively regulated TLR4-induced NF-κB signaling pathway components (14).
Many studies have confirmed the value of miRNA for immunomodulation, and some studies have also suggested that miRNA may serve as useful markers guiding etiological examinations. The most common pathogen of sepsis is Gram-negative bacteria, followed by Gram-positive bacteria (15). Not all patients with sepsis have positive blood cultures (16). A study on the specificities of miRNAs in bacterial infections during sepsis showed the serum levels of seven circulating miRNA targets (miR-133a, miR-133b, miR-122, miR-205, miR-1899, miR-714, and miR-291b) were regulated during, and Gram-positive bacterial infection. However, the serum levels of miR-16, miR-17, miR-20a, miR-26a, miR-26b, miR-106a, miR-106b, and miR-451 were selectively elevated in Gram-negative bacteria-induced sepsis (17). miRNA also supplies a new basis for the early etiological treatment of sepsis.
The high-throughput sequencing of miRNA is an experimental technique at the gene level. It sequences the genome to obtain the miRNA tags and analyze and identify the existing miRNAs, known miRNAs, and novel miRNAs; from the expressions of miRNAs in different samples, it obtains the expression profile of all miRNAs, which can be used for searching for the possible candidate genes that cause a specific disease. This technique has been widely applied in clinical studies. For instance, Luna et al. used this technology to study the candidate miRNA for hepatocellular carcinoma (HCC) aside from chronic hepatitis c virus infection (18); Nowakowski et al. used this technology to explore miRNA as a marker gene that controls the rate of glial proliferation during brain development (19).
The studies mentioned above suggest that serum miRNAs are involved in a series of pathophysiological processes of many diseases, including sepsis. So, can miRNAs be potential markers of sepsis? Some earlier articles have proposed using miRNAs as biomarkers of sepsis. For example, studies have found the low expressions of miR-223 and miR-146a might be a diagnostic criterion of sepsis (20,21). However, to our knowledge, no literature has elucidated the miRNAs roles in sepsis pathogenesis and the assessment of its severity and prognosis. Therefore, a deeper understanding of the potentially differential expressions of miRNAs in patients with sepsis and patients with septic shock and the possible correlations between miRNAs and clinical features/laboratory findings of sepsis would be highly helpful for further studies on the development and severity of sepsis and the early interventions of this disease. The objectives of our current study were to explore the difference of serum miRNA expressions in sepsis patients by using high-throughput sequencing technology and analyze whether miRNAs are potential biomarkers for estimating the severity and prognosis of sepsis; also, we explored the pathogenesis, treatment, and prognosis of sepsis at the molecular level.
Our current study can be divided into two parts:
- Serum miRNAs were extracted in septic patients and healthy individuals by using ultracentrifugation. The differential expression of miRNAs in the serum was detected by high-throughput sequencing technology. The differential expressions of miRNAs in these two groups were compared, and the differentially expressed candidate gene miR-206 was screened. The expressions of miR-206 in these two groups were verified by qRT-PCR.
- Serum miR-206 expression was detected in patients with sepsis and those with septic shock by using qRT-PCR. These patients were divided into the miR-206 low-expression group and miR-206 high-expression group by using a quartile method according to miR-206 expression level. The clinical data, laboratory findings, and prognosis were compared between these two groups. The correlations between miR-206 and disease evaluation indicators were analyzed to identify whether serum miR-206 level can be used as a biomarker for the early assessment of sepsis severity and prognosis and whether it is valuable in predicting the progression and outcome of sepsis.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/apm-20-1391).
Methods
Subjects
Healthy individuals (n=28) were randomly selected from the health check-up center of Affiliated Hospital of Nantong University. The blood samples of sepsis (n=63) and septic shock (n=30) were collected from 93 inpatients in the Emergency Intensive Care Unit (EICU) of the Affiliated Hospital of Nantong University during the period from September 2018 to January 2020. The diagnosis of sepsis or septic shock was for the International Guidelines for Management of Sepsis and Septic Shock: 2016. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and has been approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Approval number: 2018-L062), and all the subjects and their families signed the informed consent documents.
Inclusion criteria
Diagnostic criteria for sepsis
According to the International Guidelines for Management of Sepsis and Septic Shock: 2016, the Sequential (Sepsis-related) Organ Failure Assessment (SOFA) score should be ≥2 (Table 1).

Full table
Diagnostic criteria for septic shock
Exclusion criteria
The exclusion criteria included: (I) younger than 18 years; (II) complicated by multiple organ dysfunction caused by other non-infectious factors; (III) complicated by an autoimmune disease treated with steroids or immunosuppressive therapy; (IV) complicated by other diseases that affect blood coagulation; (V) with malignant tumors; (VI) complicated by other diseases that may affect the results of this study.
Collection of clinical data
Results of routine blood tests, biochemical indicators, and serological parameters, including hs-CRP, PCT, ESR, and blood cultures, were provided by the Clinical Laboratory of the Affiliated Hospital of Nantong University.
Experiments
Sample collection
- Peripheral venous blood sample (about 3 mL) was collected using an EDTA-containing tube. Store whole blood at 4 degrees Celcius (or in an insulated container with ice).
- The sample was centrifuged at 3,000 rpm at 4 °C for 10 minutes;
- The supernatant was pipetted into a 1.5-mL enzyme-free centrifuge tube and centrifuged at 13,000 rpm for 2 minutes;
- The supernatant was pipetted into a 1.5-mL conical bottom centrifuge tube, and the precipitate was discarded; and
- The sample was stored in a −80 °C refrigerator for subsequent experiments.
Detection of the differential expressions of miRNAs in the serum by high-throughput sequencing technology
- The serum supernatant specimens were separately extracted from 3 septic patients and 3 healthy individuals;
- Total RNA was extracted from specimens with Trizol reagent;
- Gel electrophoresis separated DNA fragments (18–30 nt);
- RNA adaptors for 5'- and 3'-ends are ligated to the purified small RNAs for use in reverse transcription and PCR amplification;
- About 140 bp of the fragment was recovered and purified by agarose gel electrophoresis;
- Sample quality control (QC) of the nucleic acid was performed with qPCR on the Agilent 2100 bioanalyzer. The sample was a load for sequencing, to show the tag sequences of small RNA in the sample and identify the miRNAs; and
- miRNAs with expressions changed by more than 2-fold, as determined by high-throughput sequencing, were used as candidate genes for prediction of miRNA target genes and functional enrichment analysis.
Total RNA extraction from serum
- The reagents were pipetted to an RNase-free 1.5-mL centrifuge tube in the following order (Table 2);
- The tubes were placed in the water bath (56 °C) for 10 minutes;
- Add 1,000 µL of absolute ethanol and mix thoroughly by inverting the tube;
- Put the adsorption column into the collection tube, pipette 760 µL of the mixture into the adsorption column. The mixture was left still for 2 min and then centrifuged at 12,000 rpm at 4 °C for 1 min. Discard the waste liquid;
- The remaining 760 µL of the solution was pipetted into the adsorption column, and the above steps were repeated;
- The wash solution A was prepared as follows (Table 3);
- The wash solution B was prepared as follows (Table 4);
- Pipette 500 µL of wash solution A into the adsorption column. The mixture centrifuged at 12,000 rpm at 4 °C for 1 min. Discard the waste liquid;
- Pipette 500 µL of wash solution B into the adsorption column. The mixture centrifuged at 12,000 rpm at 4 °C for 1min. Discard the waste liquid;
- The mixture was centrifuged at 12,000 rpm at 4 °C for 2 minutes to remove the residual wash solution;
- Pipette 30 µL of the elution solution to a new RNase-free 1.5-mL centrifuge tube and let stand for 3 minutes. The mixture was centrifuged at 12,000 rpm at 4 °C for 2 minutes, and finally, the RNA stock solution was collected;
- The RNA concentration was determined by using a protein/nucleic acid quantifier;
- The RNA stock solution was stored in a −80 °C refrigerator for subsequent experiments.

Full table

Full table

Full table
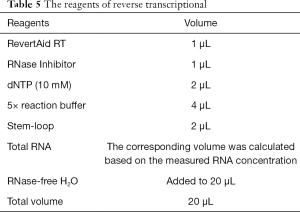
Reverse transcription
- The following reagents were added following the steps required by the reverse transcription kit (Thermo Fisher Scientific, USA). The total reaction volume was 20 µL (Table 5);
- The conditions for reverse transcription reaction were as follows: 42 °C for 60 min, 72 °C for 60 min, and 4 °C for constant temperature, one cycle;
- Store the reverse-transcribed cDNA in the refrigerator at −20 or −80 °C.
Full table
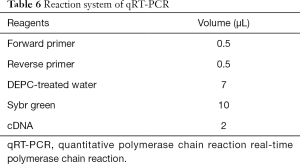
qRT-PCR
- Reaction system (Table 6);
- Amplify according to miRNA parameters.
Full table
Statistical analysis
Statistical analysis was performed using SPSS 25.0 (SPSS Inc., Chicago, IL, USA) software package. Two-sided P values were designed to test hypotheses, and a P value of <0.05 was considered statistically significant.
- Normally distributed measurement data are expressed as mean ± standard deviation (‾x ± SD), and non-normally distributed measurement data are presented as medians (interquartile ranges). Frequencies express Count data (percentages).
- Normally distributed measurement data are compared using independent sample t-test, non-normally distributed measurement data are compared using the Mann-Whitney U test, and count data are analyzed using a chi-square test.
- Correlation analysis was based using Pearson correlation analysis.
Results
Differential expressions of miRNAs in the serum measured by high-throughput sequencing technology
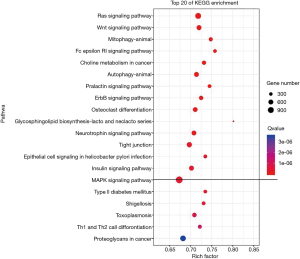
Serum miRNA expressions in septic patients and healthy individuals were determined by high-throughput sequencing technology. Compared with healthy individuals, 29 miRNA molecules were down-regulated, and 25 molecules were up-regulated in the serum samples of septic patients (Figure 1). GO, and KEGG enrichment analyses showed that MAPK and some other pathways were involved (Figure 2). miR-206, which was up-regulated and had an expression changed by more than 2-fold, was screened as a candidate gene.
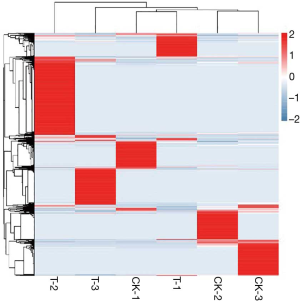
Verification of the differentially expressed molecule miR-206 with qRT-PCR
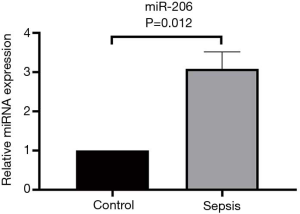
The miR-206 with differential expression of more than 2 times in high-throughput sequencing was screened, and the serum samples of 10 healthy individuals and 10 septic patients were collected. QRT-PCR amplified the sample size to verify the serum miR-206 expression in both groups. It was found that serum miR-206 expression was significantly higher in septic patients than in healthy individuals (P=0.012) (Figure 3).
Analysis of miR-206 expressions in sepsis and septic shock with qRT-PCR
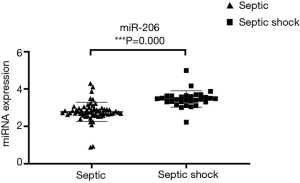
The serum miR-206 expression in the two groups was compared. It was found that the serum miR-206 expression was significantly higher in patients with septic shock than in patients with sepsis (P<0.05) (Figure 4).
Baseline data of the subjects
Serological findings
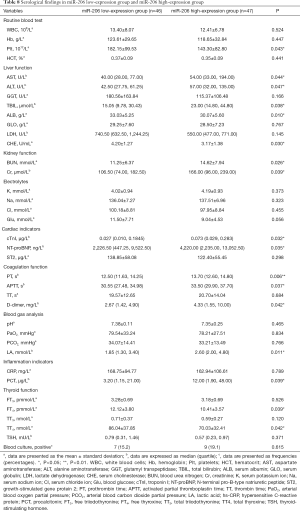
Baseline datas including gender, age, and length of EICU stay showed no significant difference (all P>0.05 Table 7). Serological findings differed between the miR-206 low-expression group and miR-206 high-expression group. The platelet count and serum albumin, cholinesterase, free T4, and thyroxine levels were significantly lower in the miR-206 high-expression group than in the miR-206 low-expression group. Compared with the miR-206 low-expressed group, the miR-206 high-expressed group had significantly higher levels of aspartate aminotransferase, alanine aminotransferase, total bilirubin, urea nitrogen, blood creatinine, troponin I, NT-proBNP, D-dimer, lactate, and PCT and significantly longer prothrombin time. It activated partial thromboplastin time (all P<0.05). Other indicators, including white blood cell count, hemoglobin, electrolytes, and blood cultures, showed no significant difference between these two groups (all P>0.05) (Table 8).

Full table
Full table
SOFA score and APACHE-II score
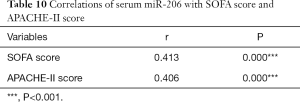
SOFA scores and APACHE-II scores were significantly higher in the miR-206 high-expression group than in the miR-206 low-expression group. Pearson correlation analysis showed that miR-206 was positively correlated with SOFA score and APACHE-II score (both P<0.05) (Tables 9 and 10).

Full table
Full table
Prognosis and outcomes
The prognosis and outcomes significantly differed between the miR-206 low-expression group and the miR-206 high-expression group. The in-hospital mortality rate was significantly higher in the miR-206 high expression group than in the miR-206 low-expression group (P=0.014) (Table 11).

Full table
Discussion
The in-hospital mortality rate is about 10% in patients with sepsis but can reach 40% in patients with septic shock. Therefore, accurate assessment of the severity and prognosis of sepsis, especially septic shock, is important for its tailored treatment. At present, serological tests, SOFA scores, and APACHE-II scores are the common clinical indicators for assessing the pathogenesis and prognosis of sepsis (22); yet, the diagnosis of septic shock is on the arterial pressure and serum lactate level after adequate volume resuscitation and persistent hypotension with vasopressors. Since the pathogenesis of sepsis is intricate, the above indicators cannot accurately and intuitively reflect the clinical features of sepsis, nor can they be used as a sensitive and specific indicator for predicting sepsis severity and prognosis in its early stage. miRNAs are widely involved in gene regulation of various links in the human body. As key gene expression-regulating molecules, they play critical roles in the pathogenesis and pathophysiological processes of various diseases. Their expressions vary in different diseases and are also closely related to the state of a specific disease (23). miRNA can be detected in various tissues and body fluids, which sheds new light on studies on various diseases at the miRNA level (24). Since circulating miRNAs are easy to obtain without severe damage, the potential molecular mechanisms of serum miRNA over-expression or inhibition in various diseases have become popular research topics. Serum miRNA expression is superior to traditional biological markers in terms of sensitivity and specificity.
Many studies have proved that miRNAs are involved in the inflammatory response and parenchymal cell apoptosis and targeted regulate inflammation-related signaling pathways (25). As obvious immune dysregulation exists in the clinical course of sepsis (26), miRNAs must have also been widely involved in the regulation of sepsis. Accordingly, animal experiments and clinical studies on miRNAs in the context of sepsis have also been carried out. The objectives of our current study were to investigate whether miRNAs can be used as potential predictive markers of sepsis and whether miRNAs are correlated with the severity and prognosis of sepsis. Serum miRNA expressions in septic patients and healthy individuals were determined by high-throughput sequencing technology; compared with healthy individuals. The septic patients had 29 down-regulated miRNA molecules and 25 up-regulated molecules. miR-206, which was up-regulated and had an expression changed by more than 2-fold, was screened as a candidate gene. The serum expression of miR-206 in septic patients and healthy individuals were verified by qRT-PCR, which proved that the serum miR-206 expression was up-regulated in septic patients. Also, qRT-PCR revealed that serum miRNA-206 expression was significantly higher in patients with septic shock than in patients with sepsis (P=0.000). These patients were divided into a miR-206 low-expressed group and miR-206 high-expressed group by using a quartile method according to miR-206 expression level. The statistical analysis showed platelet count, serum albumin, serum cholinesterase, free T4, thyroxine, aspartate aminotransferase, alanine aminotransferase, total bilirubin, urea nitrogen, blood creatinine, troponin I, N-terminal pro-B-type natriuretic peptide, prothrombin time, activated partial thromboplastin time, D-dimer, lactic acid, and procalcitonin significantly differed between these two groups. Therefore, miRNA-206 is correlated with the severity of sepsis and may be used as a predictor of sepsis severity.
APACHE-II score and SOFA score have been widely applied for assessing the severity and prognosis of sepsis (27). However, both covered various items and have limited values in assessing sepsis severity and prognosis in the early stages. At present, the relationships of serum miRNAs with SOFA score and APACHE-II score remain unclear, and the regulatory role and prognostic value of miRNA in sepsis are poorly understood. In our current study, there were significant differences in the SOFA score, APACHE-II score, and hospital mortality between the miR-206 low-expression group and the miR-206 high-expression group. The SOFA score, APACHE-II score, and in-hospital mortality of miR-206 high-expression group were significantly higher than those of the miR-206 low-expression group, and Pearson correlation analysis showed that serum miR-206 was positively correlated with SOFA score and APACHEII score. Thus, miR-206 may be related to the severity and prognosis of sepsis, and its specific regulatory role warrants further investigations.
Since miRNAs are negative regulators of target genes related to cell proliferation, apoptosis, and migration, most of the previous studies have focused on their roles as oncogenes or tumor suppressors (28). Therefore, the role of miR-206 in the growth, migration, and invasiveness of tumors (e.g., lung cancer and prostate cancer) has been explored in recent years. Experiments have confirmed that miRNA-206 is a member of the muscle-specific miRNA family. These miRNAs are known as muscle groups and play an important role in the formation and development of cardiac and skeletal muscles (29). It has been found that serum miR-206 regulates the barrier function of ATII cells in sepsis-related acute lung injury by targeting the Cx43 gene with unknown mechanisms. Similarly, the mechanisms via which serum miR-206 target-regulates the pathogenesis and prognosis of sepsis is still unclear.
Its small sample size limited our current study and non-multicenter design. In our future studies, we will further increase the sample size to comprehensively and accurately analyze the value of serum miR-206 in assessing the severity and prognosis of sepsis. Our future research topics will also address many problems. For example, there is evidence that miRNAs are transported from microvesicles and exosomes to the target sites during infection (30); so, are miRNAs in the serum of patients with sepsis also transported through the microvesicles and exosomes? How does it work? Is there any difference in the expression levels of miRNAs between serum and exosomes in septic patients? Do they play different regulatory roles in the disease process? Although miRNAs may provide new possibilities for the diagnosis, prognosis, and treatment of diseases, there are still many problems to be solved. However, new research advances will ultimately reveal the molecular mechanisms of many diseases, and the medical problems that seriously threaten human health will eventually be overcome.
In summary, in our pilot experiment, we analyzed the differentially expressed miRNAs in the serum samples of septic patients and healthy individuals and screened the up-regulated miR-206 in septic patients. The subsequent experiment revealed that there was a difference in serum miR-206 expression between patients with sepsis and those with septic shock, and miR-206 was highly expressed in patients with septic shock. Patients with sepsis or septic shock were divided into miR-206 low-expressed group and miR-206 high-expressed group by using a quartile method according to miR-206 expression level. Compared with the miR-206 low-expressed group, the miR-206 high-expressed group had significantly higher levels of aspartate aminotransferase, alanine aminotransferase, total bilirubin, urea nitrogen, blood creatinine, troponin I, NT-proBNP, D-dimer, lactate, and procalcitonin, significantly longer prothrombin time and activated partial thromboplastin time, and significantly higher SOFA score, APACHE-II score, and in-hospital mortality rate. The platelet count, serum albumin, serum cholinesterase, free T4, and thyroxine levels were significantly lower in the miR-206 high-expression group than in the miR-206 low-expression group. Serum miR-206 was positively correlated with the SOFA score and APACHE-II score. Therefore, miR-206 has a certain correlation with the severity and prognosis of sepsis and may serve as a potential biomarker for sepsis. However, the exact mechanism via which miR-206 targeted regulates the development and progression of sepsis is still unclear and calls for further investigations.
Conclusions
With patients with sepsis or septic shock as the main subjects, we investigated the serum expression of miR-206 in patients with sepsis and found serum miR-206 expression is positively correlated with the severity and prognosis of sepsis. Thus, it may serve as a potential biomarker for assessing the severity and prognosis of sepsis. This study provides a theoretical basis for serum miRNA as a predictive indicator for the occurrence, development, and prognosis of sepsis, and testing of miR-206 may be a simple tool that can accurately reflect the clinical features of sepsis.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China Youth (81801893) and the Nantong Clinical Medicine Research Center (HS2019005).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-1391
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-1391
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-1391). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The diagnosis of sepsis or septic shock was for the International Guidelines for Management of Sepsis and Septic Shock: 2016. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and has been approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Approval number: 2018-L062), and all the subjects and their families signed the informed consent documents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Uhel F, Peters-Sengers H, van der Poll T. Initiation of renal replacement therapy in patients with sepsis: more to it than meets the eye. Ann Transl Med 2018;6:S130. [Crossref] [PubMed]
- Lin X, Luo H, Yan X, et al. Interleukin-34 ameliorates survival and bacterial clearance in polymicrobial sepsis. Crit Care Med 2018;46:e584-90. [Crossref] [PubMed]
- Giza DE, Fuentes-Mattei E, Bullock MD, et al. Cellular and viral microRNAs in sepsis: mechanisms of action and clinical applications. Cell Death Differ 2016;23:1906-18. [Crossref] [PubMed]
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297-325. [Crossref] [PubMed]
- Correia CN, Nalpas NC, McLoughlin KE, et al. Circulating microRNAs as Potential Biomarkers of Infectious Disease. Front Immunol 2017;8:118. [Crossref] [PubMed]
- Lavrentieva A, Papadopoulou S, Kioumis J, et al. PCT as a diagnostic and prognostic tool in burn patients. Whether time course has a role in monitoring sepsis treatment. Burns 2012;38:356-63. [Crossref] [PubMed]
- Yan HX, Du J, Fu J, et al. Microarray-based differential expression profiling of long noncoding RNAs and messenger RNAs in formalin-fixed paraffin-embedded human papillary thyroid carcinoma samples. Transl Cancer Res 2019;8:439-451. [Crossref]
- Kingsley SMK, Bhat BV. Role of microRNAs in sepsis. Inflamm Res 2017;66:553-69. [Crossref] [PubMed]
- Ma Y, Liu Y, Hou H, et al. MiR-150 predicts survival in patients with sepsis and inhibits LPS-induced inflammatory factors and apoptosis by targeting NF-κB1 in human umbilical vein endothelial cells. Biochem Biophys Res Commun 2018;500:828-37. [Crossref] [PubMed]
- Di A, Xiong S, Ye Z, et al. The TWIK2 Potassium Efflux Channel in Macrophages Mediates NLRP3 Inflammasome-Induced Inflammation. Immunity 2018;49:56-65.e4. [Crossref] [PubMed]
- Funahashi Y, Kato N, Masuda T, et al. miR-146a targeted to splenic macrophages prevents sepsis-induced multiple organ injury. Lab Invest 2019;99:1130-42. [Crossref] [PubMed]
- Zhang Z, Liang K, Zou G, et al. Inhibition of miR-155 attenuates abdominal aortic aneurysm in mice by regulating macrophage-mediated inflammation. Biosci Rep 2018;38:BSR20171432. [Crossref] [PubMed]
- Essandoh K, Li YT, Huo JZ, et al. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 2016;46:122-31. [Crossref] [PubMed]
- Zhang H, Li H, Shaikh A, et al. Inhibition of MicroRNA-23b Attenuates Immunosuppression During Late Sepsis Through NIK, TRAF1, and XIAP. J Infect Dis 2018;218:300-11. [Crossref] [PubMed]
- Dombrovskiy VY, Martin AA, Sunderram J, et al. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: a trend analysis from 1993 to 2003. Crit Care Med 2007;35:1244-50. [Crossref] [PubMed]
- Abram TJ, Cherukury H, Ou CY, et al. Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR. Lab Chip 2020;20:477-89. [Crossref] [PubMed]
- Wu SC, Yang JC, Rau CS, et al. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS One 2013;8:e77936. [Crossref] [PubMed]
- Luna JM, Scheel TK, Danino T, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell 2015;160:1099-110. [Crossref] [PubMed]
- Nowakowski TJ, Rani N, Golkaram M, et al. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci 2018;21:1784-92. [Crossref] [PubMed]
- Wang H, Zhang P, Chen W, et al. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS One 2012;7:e38885. [Crossref] [PubMed]
- Quinn EM, Wang JH, O'Callaghan G, et al. MicroRNA-146a is upregulated by and negatively regulates TLR2 signaling. PLoS One 2013;8:e62232. [Crossref] [PubMed]
- Mustafić S, Brkić S, Prnjavorac B, et al. Diagnostic and prognostic value of procalcitonin in patients with sepsis. Med Glas (Zenica) 2018;15:93-100. [PubMed]
- Liu H, Lei C, He Q, Pan Z, et al. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer 2018;17:64. [Crossref] [PubMed]
- Fejes Z, Szilágyi B, Kappelmayer J, et al. Alteration in the expression of platelet microRNAs in diseases with abnormal platelet activation. Orv Hetil 2018;159:1962-70. [Crossref] [PubMed]
- O'Neill LA, Sheedy FJ, McCoy CE. MicroRNAs:the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 2011;11:163-75. [Crossref] [PubMed]
- Yarema TC, Yost S. Low-dose corticosteroids to treat septic shock:a critical literature review. Crit Care Nurse 2011;31:16-26. [Crossref] [PubMed]
- Szederjesi J, Almasy E, Lazar A, et al. An Evaluation of Serum Procalcitonin and C-Reactive Protein Levels as Diagnostic and Prognostic Biomarkers of Severe Sepsis. J Crit Care Med (Targu Mures) 2015;1:147-53. [Crossref] [PubMed]
- Moreno EC, Pascual A, Prieto-Cuadra D, et al. Novel Molecular Characterization of Colorectal Primary Tumors Based on miRNAs. Cancers (Basel) 2019;11:346. [Crossref] [PubMed]
- McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 2008;1779:682-91. [Crossref] [PubMed]
- Mori MA, Ludwig RG, Garcia-Martin R, et al. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab 2019;30:656-73. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)