
Microbial quantitation of colostrum from healthy breastfeeding women and milk from mastitis patients
Introduction
Human breast milk is a resource of microorganisms, which can colonize in the infant intestine to improve the microflora establishment (1). However, the amounts of microbiota in the breast milk fluctuate during the lactation and vary depending on the postpartum time (2).
Colostrum, the breast milk generated within 5 or 6 days postpartum (3), contains a relatively large amount of microbiota which colonizes in mammary gland during the third trimester of pregnancy (2,4). Culture-dependent and -independent analyses of bacteria have revealed that the colostrum is a source of Staphylococcus, Streptococcus, Lactobacillus, Bifidobacterium and Enterobacterium (5-7). These opportunistic pathogens and probiotics are in a dynamic balance and important for the maintenance of mammary homeostasis (8,9). However, available studies on the microbial quantitation of colostrum focus are from America and Europe, and little is known about the quantitation of microorganism in the colostrum of Chinese women. In addition, the presence of lactational mastitis may also affect the amounts of microbiota in the breast milk.
Lactational mastitis is the most common disease during lactation and usually causes the earlier weaning or breast appearance damage. Lactational mastitis is closely related to the mammary bacterial imbalance in which the microbiota responsible for mastitis increase while normal mammary microbiota decreases (10,11).
Specifically, Staphylococcus is the most common microorganism causing mastitis (12), and Streptococcus and Escherichia are less common in the mastitis (13). Besides, small amounts of probiotics such as Bifidobacterium and Lactobacillus can also colonize in the mammary gland (14). However, few studies have conducted to investigate the distribution of microorganisms in the breast of Chinese women.
This study was to quantify the microbiota in bilateral breast milk from healthy breastfeeding women and lactational mastitis patients in China, and investigate the distribution of microorganism, aiming to provide evidence on the pathogenesis of lactational mastitis. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/apm-20-56).
Methods
Study design
The healthy Chinese breastfeeding women from the Shanghai Maternity and Infant Care Centre of Putuo District, China and the Chinese women with lactational mastitis from the Shuguang Hospital of Shanghai University of Traditional Chinese Medicine, China were included into present study between January 2017 and December 2017. The study was approved by the Institutional Review Board (IRB) of Shuguang Hospital (2017-551-34-01) and Ethics Committee of Shanghai Maternity and Infant Care Centre in Putuo District (2017-3). This study was conducted in accordance with the Chinese Measures for the Ethical Review of Biomedical Research Involving Humans, the Declaration of Helsinki and International Ethical Guidelines Biomedical Research Involving Human Subjects. Informed consent was obtained from all subjects before study. And the sample size was estimated based on the comparison of the mean value of two independent samples.
The inclusion criteria for the healthy breastfeeding women were as follows: (I) they were healthy and breast-feeding; (II) written informed consent was obtained; and (III) postpartum colostrum was collected within 5 days after delivery. Exclusion criteria were as follows: (I) there was evident maternal illness; and (II) there was usage of antibiotics during lactation.
The inclusion criteria for mastitis patients were as follows: (I) they were puerperal or lactational women, (II) they had unilateral breast lump, (III) there were local redness and pain; (IV) women had or had no fever or fatigue. The exclusion criteria were as follows: (I) there was use of antibiotic treatment, (II) there was mammary abscesses, (III) there was Raynaud’s syndrome, (IV) there was any other mammary pathology.
Collection and processing of milk samples
The colostrum samples were collected from healthy breastfeeding women within 5 days after delivery, and the breast milk samples were collected from the mastitis women at the time of hospital visit. 5 mL of milk was collected from each side of the breast.
Before the milk collection, the areola and nipple area were sterilized by using 75% alcohol. The first drop of milk (about 150 µL) was discarded to avoid contamination. Samples were collected from bilateral breasts into sterile containers and kept at 4 °C immediately until analysis. Milk was cultured within 2 h after collection, and bacterial DNA was extracted within 24 h after collection. The remaining samples were stored at −80 °C.
Milk culture and bacterial identification
The specific bacteria were detected by culturing of milk from mastitis patients. Milk was spread on the Nutrient agar and Mac Conkey agar (Yihua Medical Technology, Shanghai, China) and incubated for 48 h at 37 °C. The number of colonies on the agar was determined by using the VITEK 2 Compact automatic bacterial identification analyzer (VK2C 6030, BIOMERIEUX, U.S.).
Bacterial DNA extraction
Total bacterial DNA was extracted from the colostrum and mastitis milk samples using a bacterial DNA Extraction Kit (TIANGEN, Beijing, China). Briefly, the milk samples (1 mL) were centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants with the fat and whey layer were removed, and the pellet bacteria were processed for the extraction of bacterial DNA following the manufacturer’s instructions. The protocol included an initial rupture of the bacterial wall by 30-min incubation with lysozyme (20 mg/mL) at 37 °C.
QPCR amplification
Preparation of standards
The qPCR was done with bacterial genomic DNA PCR products. Briefly, the PCR products of each primer pair were purified and cloned into EcoRV of pUCS vector and transduced into Top 10 strain. Cloned standard plasmid was confirmed by sequencing. For PCR quantification, four plasmids were purified and the concentration was adjusted to 108, 107, 106, 105, 104, 103 and 102 copies/mL standard solution. PCR standards were added, and the standard curve was obtained.
Quantitation of colostrum/mastitis milk DNA
Microbial quantitation was based on the conserved marker gene 16s rRNA for bacterial genus including Bifidobacterium, Lactobacillus, Staphylococcus and Streptococcus.
Real-time PCR quantitation for target bacterial gene in both colostrum and mastitis milk was conducted with ABI 7900HT Fast Real-time PCR System (Applied Biosystems, ThermoFisher, U.S.). The reactions were carried out in a 25 µL mixture containing 12.5 µL of SYBR Green Master Mix (2×), 0.1 µL of each SP/ASP (100 µM), 5.0 µL of template (1 ng/µL) and 7.3 µL of double distilled water. The primers were used to amplify the V3 region of bacterial 16s rDNA (Table 1). The PCR conditions used were as follows: 2 min at 95 °C, 40 cycles of 95 °C for 15 s, 60 °C for 45 s, followed by 95 °C for 15 s, 60 °C for 15 s, and 95 °C for 15 s
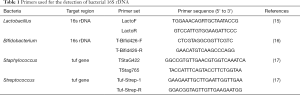
Full table
Analysis of cytokines in breast milk
The quantities of CRP, Sodium, TNF-α, IL-1, IL-6 and IL-8 in both colostrum and mastitis milk were determined additionally. The C-reactive protein and sodium concentration in the breast milk were measured using the High sensitivity C-reactive protein determination Kit (Beckman Coulter, USA) and Electrolytes detection kit (Beckman Coulter, USA) on a fully automated Chemiluminescence instrument (Beckman Coulter, AU5800, USA). The cytokines in the breast milk samples were testing using a Aim-Plex Human Cytokine Plex Panel kit (Tianjin Kuang Bo Tongsheng Biotechnology Co., Ltd, China) and analyzed through the Cellquest analysis platform (BD Calibur, USA). The analyses were performed according to the manufacturer’s instructions.
Statistical analysis
The statistical analysis was done with IBM SPSS version 21.0 (Chicago, IL, U.S.) and Graphpad Prism version 5.0 (La Jolla, CA, U.S.). Categorical variables were compared using the Chi-square test or Fisher’s exact test. Continuous variables were compared using student’s t test, one-way analysis of variance, or Mann-Whitney U test. Collections between the bacterial generic quantity were evaluated by Pearson’s correlation coefficient. A value of two-tailed P<0.05 was considered statistically significant.
Results
Clinical characteristics of healthy breastfeeding and mastitis women
A total of 190 breastfeeding women were recruited into present study: 104 healthy breastfeeding women and 86 breastfeeding women diagnosed with lactational mastitis. There were no marked differences between healthy women and mastitis women in the maternal age, gestational age, delivery mode and parity (Table 2).
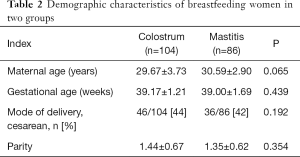
Full table
The milk samples from healthy breastfeeding women were collected at 0–5 days after delivery. The distribution of postpartum time is shown in Table 3. According to the classification of human breast milk (18), the milk collected from healthy breastfeeding women in the present study was the colostrum.
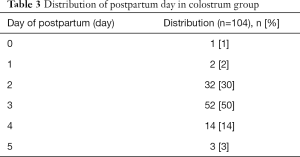
Full table
Quantitation of microbiota in colostrum
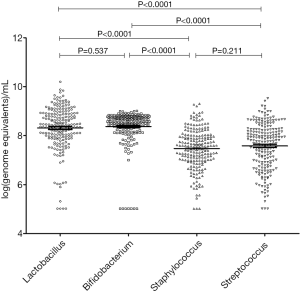
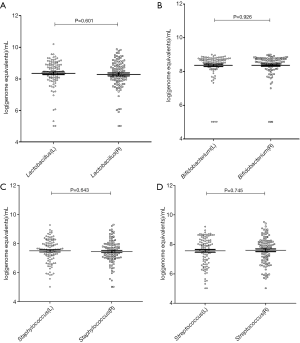
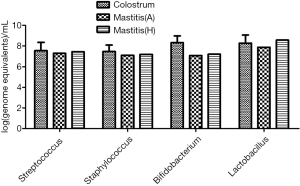
Real-time PCR was successfully performed in the colostrum samples from 104 healthy breastfeeding women, and the quantity of Lactobacillus, Bifidobacterium, Staphylococcus and Streptococcus at genus level was detected. Among these bacteria, the amounts of Lactobacillus and Bifidobacterium were significantly higher than those of Staphylococcus and Streptococcus (P<0.0001). The amount of Lactobacillus was similar to that of Bifidobacterium, and the amount was also comparable between Staphylococcus and Streptococcus (P>0.05) (Figure 1). Furthermore, the amount of each bacterium was also similar between bilateral breasts (P>0.05) (Figure 2). There was a correlation between the quantity of any two bacteria (P<0.0001) (Table 4).

Full table
Microbial identification and quantitation in the mastitis milk
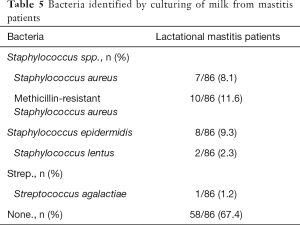
Real-time PCR was also done to detect the bacterial DNA in the bilateral breast milk from 86 lactational mastitis patients. The quantity of Lactobacillus, Bifidobacterium, Staphylococcus and Streptococcus at genus level was detected. In addition, the bacterial culture was performed with the milk from the affected breast. Interestingly, bacteria were not identified in the majority of samples (58/86). The dominant pathogens isolated were Staphylococcus spp. including S.A. (Staphylococcus aureus) (7/86), M.R.S.A. (Methicillin-resistant Staphylococcus aureus) (10/86), S.E. (Staphylococcus epidermidis) (8/86) and S.L. (Staphylococcus lentus) (2/86). S. agalactiae (Streptococcus agalactiae), a less common pathogen in the human mastitis (19), was isolated from 1 patient (Table 5). Furthermore, qPCR indicated that the amounts of four detected bacteria (except for Bifidobacterium) in bilateral milk samples were similar in all patients with unilateral mammary lesion (P>0.05) (Figure 3).
Full table
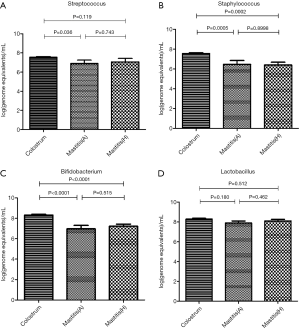
Microbiota quantitation of the colostrum and mastitis milk
Based on the above findings, the mastitis could be divided into strong virulent bacteria (Staphylococcus aureus, M.R.S.A.)-mediated mastitis, weak virulent bacteria (Staphylococcus vulgaris, Streptococcus mitis)-mediated mastitis, Streptococcus agalactiae mediated mastitis and bacteria-unrelated mastitis. The amounts of four detected bacteria were different from that of the colostrum. This finding was different from that in the comparison between mastitis and healthy women.
The amounts of four detected bacteria in the colostrum were significantly higher than those in the healthy breast and even in the affected breast of mastitis patients (P<0.01) (Figure 3).
In 17 patients with mastitis caused by S.A. or M.R.S.A (the most common pathogen in mastitis), the amount of Staphylococcus in the colostrum was significantly lower than in the milk from affected breast, but similar to that in the milk from unaffected breast (Figures 4,5). In 10 patients with mastitis caused by S.E. and S.L, the amount of Staphylococcus in the colostrum was similar to or higher than that in the milk from affected or unaffected breast (Figures 6,7). In the Staphylococcus-related mastitis patients, the amount of Bifidobacterium in the colostrum was significantly higher than in the milk from affected or unaffected breast (P<0.01) (Figures 4-7). However, the amount of Lactobacillus was similar among the colostrum, milk from affected breast and milk from unaffected breast (P>0.05) (Figures 4-7).
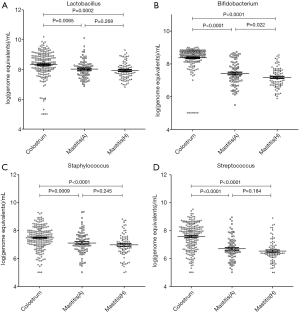
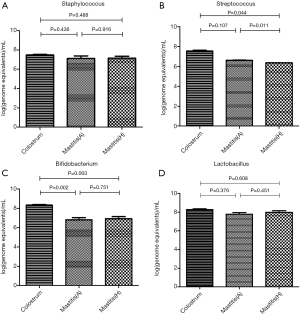
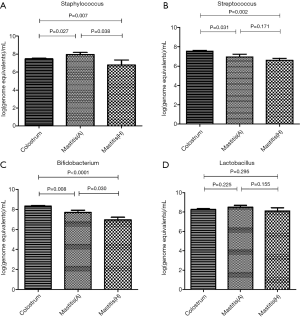
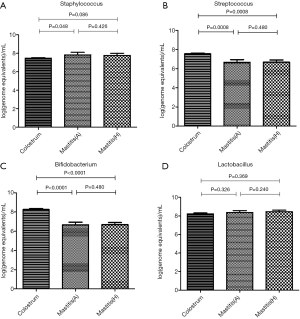
The patients with bacteria unrelated mastitis accounted for 67.4% (Table 5). In these cases, the abundance of four detected bacteria in the colostrum was significantly higher than that in the milk from affected or unaffected breast (P<0.01) (Figure 8).
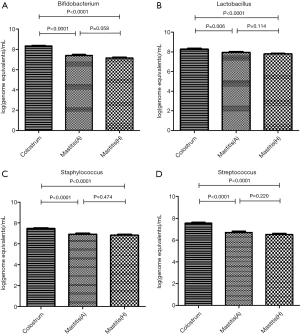
In only one patient, S. agalactiae was isolated from the milk of affected breast. The amounts of Bifidobacterium and Lactobacillus in the colostrum were slightly higher than in the milk from affected breast (P>0.05; Figure 9).
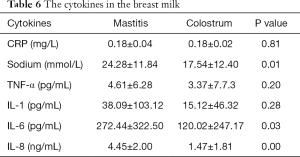
Cytokines in colostrum and mastitis milk
The quantities of Sodium, IL-6, IL-8 in mastitis milk were higher than those in colostrum respectively, but the quantities of CRP, TNF-α, IL-1 were not shown the statistical differences (Table 6).
Full table
Discussion
This study for the first time investigated the microbiota in the colostrum from healthy breastfeeding women and in breast milk from patients with lactational mastitis in China. In addition, the microbial quantity was also determined in bilateral colostrum and mastitis milk samples.
Colostrum is the human breast milk generating within 5–6 days after delivery and the first natural food for infants (3). In this study, the microbiota in the colostrum from bilateral breasts were investigated in 104 healthy Chinese breastfeeding women. Among the detected bacteria, the amounts of probiotics (Lactobacillus and Bifidobacterium) were significantly higher than those of opportunistic pathogens (Staphylococcus and Streptococcus) (P<0.0001), and the amounts between two probiotics or two pathogens were similar (P>0.05), although there were correlations among the amounts of all detected bacteria (P<0.0001). In previous study, the four investigated bacteria have also been identified in the colostrum in other countries (5-7,18,20). A Slovenian study also confirmed the relationships among the amounts of bacteria detected in the colostrum. As probiotics, Lactobacillus and Bifidobacterium have been frequently isolated from the colostrum and they play beneficial roles in the infant intestinal colonization, inhibition of pathogen bacteria, anti-inflammation and treatment of infectious mastitis (21-24). In contrast, Staphylococcus and Streptococcus are the opportunistic pathogens in the lactational mastitis. Consequently, the balance of microorganisms including probiotics and potentially infectious microorganisms are in sufficient numbers to colonize the infant intestine but do not induce an inflammatory response in the mammary ducts (25).
Moreover, this study for the first time reported the amounts of four bacteria were similar in colostrum from bilateral breasts. It may be explained as that the human breast is a symmetrical organ, and similar amounts of mammary epithelium differentiate into lactocytes with the capacity to synthesize breast milk under the action of the same hormone level (26).
In addition, the microbiota was also quantified in the milk from bilateral breasts of 86 Chinese women who were diagnosed with lactational mastitis by qPCR and bacterial culture. Mastitis is the most common disease during lactation. The main pathogen of mastitis is Staphylococcus spp. This study showed 67.4% of mastitis patients had no detectable bacteria. Thus, the amount of each bacteria investigated was comparable between the affected breast and unaffected breast, although all the mastitis patients had unilateral breast problems. The specific reason is still unclear, and few infectious pathogens isolated from the milk of affected breast might be related to the similar microbial quantity.
In addition, amount of two abundant bacteria were compared based on the qPCR and milk culture.
The amounts of four detected bacteria in the colostrum dramatically increased as compared to the milk from affected breast and unaffected breast. There was a higher load of bacterial load in colostrum compared with mature milk (7,27). Therefore, the number of microbiota in colostrum might be the top during the lactation. Once the population of opportunistic pathogens increasing while the commensal mammary microbiota decreasing, the mastitis might occur. The microorganisms quantitatively detected in this study are at the “genus” level and do not involve the inferior level of “species”. Therefore, it is impossible to absolutely distinguish the colonizing bacteria from the pathogenic bacteria. Besides, more than half of all the enrolled mastitis patients are lactation mastitis without bacteria, so the number of microorganisms in the milk is low.
However, there are no clinical symptoms in these breastfeeding women. It might be ascribed to the large amounts of probiotics as compared to the opportunistic pathogens in the colostrum. These commensal bacteria, which consist of large amounts of probiotics and relatively small amounts of opportunistic pathogens, may line regularly along the inner wall of mammary duct to protect the mammary gland from pathogen invasion and establish the ecological balance in mammary gland (28).
Furthermore, the specific pathogen causing mastitis was compared. In the mastitis caused by S.A. or M.R.S.A., the amounts of Staphylococcus in the milk from affected breast was significantly higher than that in the colostrum. In the mastitis caused by S.E. or S.L., the number of bacteria in the milk from affected breast was similar to that in the colostrum. This suggests that the pathogenesis of mastitis depends on the virulence of bacteria because the S.A. and M.R.S.A. are highly virulent (29). They can proliferate and produce toxins causing significant inflammation in the mammary tissues; the toxins may also be rapidly absorbed and even enter the blood (1). In contrast, the S.E. and S.L. belong to Coagulase Negative Staphylococcus (CNS), which are normal inhabitants of the mammary ecosystem during lactation and live at the edge between commensalism and pathogenicity (30). Thus, CNS are not as pathogenic as S.A. or M.R.S.A. and usually regarded as opportunistic pathogens (31). They are able to form thick biofilms inside the mammary ducts, inflaming the mammary epithelium and increasingly narrowing the mammary lumen, which lead to mastitis (32).
In addition, the amount of Bifidobacterium in the colostrum was remarkably higher than in the milk from affected breast and unaffected breast in patients with mastitis caused by Staphylococcus. Moreover, the amount of Bifidobacterium in the milk from affected breast was higher than that in the milk from unaffected breast in 86 mastitis patients. However, the amount of Lactobacillus was similar between bilateral breasts of mastitis patients. The probiotics in human breast milk might originate through the entero-mammary pathway. It has been suggested that intestinal dendritic cells (DCs), which can open the tight junctions between intestinal epithelial cells, can extend dendrites outside the epithelium to sample the bacteria in the intestine directly (33) and also retain the live commensal bacteria for several days in the mesenteric lymph nodes (34). Then, through the lymphatic or blood circulation, the live commensal bacteria may finally reach the ductal cavity of mammary gland under the similar action of DCs (2). The probiotics and the opportunistic pathogens colonized the mammary gland remain a dynamic balance to maintain the mammary homeostasis (8,9), and the imbalance of any cause in the mammary microbial communities will lead to mastitis (19). In this study, results showed the amount of Staphylococcus copies increased, but the amount of Bifidobacterium reduced in the Staphylococcus-induced mastitis patients, which support the role of mammary microbiota dysbiosis in the pathogenesis of lactational mastitis.
Secondly, in the study, among 58 mastitis patients without detectable bacteria, the amount of all detected bacteria in the colostrum increased significantly as compared to the milk from the affected breast and unaffected breast. This was consistent with the finding from the comparison between total colostrum and mastitis milk samples. This suggests that the colostrum is a vehicle filled with microbiota, even compared with mastitis milk. In mastitis patients without detectable bacteria, the important role of microbiota can’t explain the development of mastitis. Bacteria may augment inflammation. However, it is clear that other factors are also important for the pathogenesis of mastitis. Studies have revealed that the disease severity is closely related to the parameters of systemic and local inflammation (such as C-reactive protein in the milk and serum and sodium concentration in the milk) (35,36). The breastfeeding women with mastitis exhibit increased serum cytokines including interleukins-1 (IL-1), IL-6, IL-8 and tumor necrosis factor α (TNF-α) (37,38). Therefore, the inflammatory mediators may result in mastitis and the specific inflammatory signaling pathways activated in the host are likely to be the key determinants of susceptibility to mastitis and severity of this disease (39).
Finally, in the Streptococcus-induced mastitis patients, the pathogen isolated and identified in this study belongs to S. agalactiae. The amounts of Streptococcus, Staphylococcus, Bifidobacterium and Lactobacillus in the colostrum were higher than in the milk from mastitis patients. However, in 86 mastitis patients, S. agalactiae was isolated from 1 patient. This strain belongs to Group B Streptococcus (GBS). It is an obligate parasite of the mammary gland in the cattle (40) and the most common cause of neonatal bacterial sepsis (41).
The main limitation of this study was the method used for microbial detection. In this study, Real-time quantitative PCR could detect both alive and dead microbiota, which could not distinguish between them. In addition, it should be noted that, in this study, the microbial comparisons just focused on colostrum and mastitis milk. In future, alive microbiota in a larger genus and species should be detected, and the healthy breast milk including the transitional and mature milk be investigated.
In conclusion, results of this study indicate that colostrum may be microbial vehicle, even compared with mastitis milk. Mammary microbial dysbiosis may lead to lactational mastitis, especially mastitis induced by infectious pathogen. In addition, other mechanism may also be involved in the pathogenesis of lactational mastitis because some mastitis patients have no detectable bacteria in the milk.
Acknowledgments
We would like to thank all the technical staff at the Department of Nursing at the Shanghai Maternal and infant care hospital in Putuo District.
Funding: This work was supported partially by the Shanghai Municipal Health Commission (2016LQ020), Shanghai Science and Technology Commission (14401971000) and Shanghai Hongkou District Health Commission (HW-1902-09, HGY-YSZK-2018-07).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-56
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-56
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-56). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the IRB of Shuguang Hospital, Shanghai University of TCM (2017-551-34-01) and Ethics Committee of Shanghai Maternity and Infant Care Centre in Putuo District (2017-3), and in accordance with the Declaration of Helsinki and International Ethical Guidelines Biomedical Research Involving Human Subjects. Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fernández L, Arroyo R, Espinosa I, et al. Probiotics for human lactational mastitis. Benef Microbes 2014;5:169-83. [Crossref] [PubMed]
- Fernández L, Langa S, Martin V, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013;69:1-10. [Crossref] [PubMed]
- Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am 2013;60:49-74. [Crossref] [PubMed]
- Cabrera-Rubio R, Collado MC, Laitinen K, et al. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 2012;96:544-51. [Crossref] [PubMed]
- Aakko J, Kumar H, Rautava S, et al. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef Microbes 2017;8:563-7. [Crossref] [PubMed]
- Toscano M, De Grandi R, Peroni DG, et al. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol 2017;17:205. [Crossref] [PubMed]
- Damaceno QS, Souza JP, Nicoli JR, et al. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob Proteins 2017;9:371-9. [Crossref] [PubMed]
- Martín R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr 2003;143:754-8. [Crossref] [PubMed]
- Jeurink PV, Van Bergenhenegouwen J, Jimenez E, et al. Human milk: a source of more life than we imagine. Benef Microbes 2013;4:17-30. [Crossref] [PubMed]
- Delgado S, Arroyo R, Martin R, et al. PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis. BMC Infect Dis 2008;8:51. [Crossref] [PubMed]
- Delgado S, Arroyo R, Jimenez E, et al. Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol 2009;9:82. [Crossref] [PubMed]
- Delgado S, Garcia P, Fernandez L, et al. Characterization of Staphylococcus aureus strains involved in human and bovine mastitis. FEMS Immunol Med Microbiol 2011;62:225-35. [Crossref] [PubMed]
- Delgado S, Arroyo R, Jiménez E, et al. Mastitis infecciosas durante la lactancia: un problema infravalorado (I). Acta Pediatr Esp 2009;67:77-84.
- Michie C, Lockie F, Lynn W. The challenge of mastitis. Arch Dis Child 2003;88:818-21. [Crossref] [PubMed]
- Byun R, Nadkarni MA, Chhour KL, et al. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol 2004;42:3128-36. [Crossref] [PubMed]
- Matsuki T, Watanabe K, Tanaka R, et al. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol 1999;65:4506-12. [Crossref] [PubMed]
- Collado MC, Delgado S, Maldonado A, et al. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol 2009;48:523-8. [Crossref] [PubMed]
- Drago L, Toscano M, De Grandi R, et al. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J 2017;11:875-84. [Crossref] [PubMed]
- Contreras GA, Rodriguez JM. Mastitis: comparative etiology and epidemiology. J Mammary Gland Biol Neoplasia 2011;16:339-56. [Crossref] [PubMed]
- Obermajer T, Lipoglavsek L, Tompa G, et al. Colostrum of healthy Slovenian mothers: microbiota composition and bacteriocin gene prevalence. PLoS One 2015;10:e0123324. [Crossref] [PubMed]
- Martín V, Maldonado-Barragán A, Moles L, et al. Sharing of bacterial strains between breast milk and infant feces. Journal of Human Lactation 2012;28:36-44. [Crossref] [PubMed]
- Maldonado-Lobón JA, Diaz-Lopez MA, Carputo R, et al. Lactobacillus fermentum CECT 5716 Reduces Staphylococcus Load in the Breastmilk of Lactating Mothers Suffering Breast Pain: A Randomized Controlled Trial. Breastfeed Med 2015;10:425-32. [Crossref] [PubMed]
- Arribas B, Garrido-Mesa N, Peran L, et al. The immunomodulatory properties of viable Lactobacillus salivarius ssp. salivarius CECT5713 are not restricted to the large intestine. Eur J Nutr 2012;51:365-74. [Crossref] [PubMed]
- Arroyo R, Martin V, Maldonado A, et al. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin Infect Dis 2010;50:1551-8. [Crossref] [PubMed]
- Boix-Amorós A, Collado MC, Mira A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front Microbiol 2016;7:492. [Crossref] [PubMed]
- Pang WW, Hartmann PE. Initiation of human lactation: secretory differentiation and secretory activation. J Mammary Gland Biol Neoplasia 2007;12:211-21. [Crossref] [PubMed]
- Solís G, de Los Reyes-Gavilan CG, Fernández N, et al. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010;16:307-10. [Crossref] [PubMed]
- Fitzstevens JL, Smith KC, Hagadorn JI, et al. Systematic Review of the Human Milk Microbiota. Nutr Clin Pract 2017;32:354-64. [Crossref] [PubMed]
- Schoenfeld EM, McKay MP. Mastitis and methicillin-resistant Staphylococcus aureus (MRSA): the calm before the storm? J Emerg Med 2010;38:e31-4. [Crossref] [PubMed]
- Otto M. Staphylococcus epidermidis—the'accidental'pathogen. Nature Reviews Microbiology 2009;7:555-67. [Crossref] [PubMed]
- Pyörälä S, Taponen S. Coagulase-negative staphylococci-emerging mastitis pathogens. Vet Microbiol 2009;134:3-8. [Crossref] [PubMed]
- Jiménez E, Delgado S, Arroyo R, et al. Infectious mastitis during lactation: an underrated condition (II). Acta Pediatrica Espanola 2009;67:125.
- Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2001;2:361-7. [Crossref] [PubMed]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004;303:1662-5. [Crossref] [PubMed]
- Fetherston CM, Lai C, Hartmann PE. Relationships between symptoms and changes in breast physiology during lactation mastitis. Breastfeeding Medicine 2006;1:136-45. [Crossref] [PubMed]
- Fetherston CM, Wells JI, Hartmann PE. Severity of mastitis symptoms as a predictor of C-reactive protein in milk and blood during lactation. Breastfeeding Medicine 2006;1:127-35. [Crossref] [PubMed]
- Zheng J, Watson AD, Kerr DE. Genome-wide expression analysis of lipopolysaccharide-induced mastitis in a mouse model. Infect Immun 2006;74:1907-15. [Crossref] [PubMed]
- Mizuno K, Hatsuno M, Aikawa K, et al. Mastitis is associated with IL-6 levels and milk fat globule size in breast milk. Journal of Human Lactation 2012;28:529-34. [Crossref] [PubMed]
- Ingman WV, Glynn DJ, Hutchinson MR. Inflammatory mediators in mastitis and lactation insufficiency. J Mammary Gland Biol Neoplasia 2014;19:161-7. [Crossref] [PubMed]
- Keefe GP. Streptococcus agalactiae mastitis: a review. Can Vet J 1997;38:429-37. [PubMed]
- Ismail AQ, Yeates D, Marciano A, et al. Cow’s milk and the emergence of group B streptococcal disease in newborn babies. Neonatology 2011;100:404-8. [Crossref] [PubMed]


