Antibiotic susceptibility and molecular analyses of clinical Enterobacter cloacae isolates in Eastern Heilongjiang Province, China
Introduction
Enterobacter cloacae are intrinsically resistant to ampicillin, amoxicillin-clavulanate, and first- and second-generation cephalosporins because producing chromosome mediated AmpC β-lactamase (1). In the case of low host immunity, dysbacteriosis, or other similar cases, respiratory, bloodstream, and surgical site infections are usually caused (2). The outbreak and spread of Enterobacter cloacae has been widely reported around the world (3), and even worse, because improper use of antibiotics, the spread of drug-resistant bacteria through various resources of the health care system, lack of appropriate guidelines for the use of antimicrobial agents, the resistance situation has gradually deteriorated. It has become a significant threat to global public health (4). The primary factors leading to β-lactam antimicrobials resistance of Enterobacter cloacae may be plasmid-mediated AmpC β-lactamases (pAmpC), plasmid-coded CTX-M family extended-spectrum β-lactamases, KPC family carbapenemases, and VIM, IMP and NDM-1 metallo-β-lactamases (5). In recent years, with the widespread use of Carbapenem antibiotics, the prevalence and spread of carbapenem-resistant Enterobacteriaceae (CRE) has become a global trend.
We conducted a retrospective study to evaluate the clinical distribution and antibiotic resistance of clinical Enterobacter cloacae isolates and the molecular analyses of CREL to promote the rational use of clinical antibiotics and to demonstrate the resistance mechanism of CREL in Eastern Heilongjiang Province, China.
Methods
Setting and isolates
The retrospective study was conducted at the First Affiliated Hospital of Jiamusi University with 1,800 beds in Eastern Heilongjiang Province, China. Three hundred forty-two strains of Enterobacter cloacae were collected from January 2014 to December 2018, all isolates were found at the species level, and routine antibiotic susceptibility tests were performed by the VITEK2 compact automated system. Unfortunately, we have been preserving CREL isolates regularly since 2016, so the earlier isolates were not preserved. Seven strains of CREL were collected from September 2016 to December 2018, and 3 added strains were collected in 2019. All of them were stored in a −80 °C refrigerator for further analysis.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of all isolates was evaluated by the VITEK2 Compact automatic system AST GN card (bioMérieux, France), supplemented by a disk diffusion method. As CREL were considered only those isolates that were confirmed as carbapenems-nonsusceptible (either of ertapenem, imipenem or meropenem) according to Clinical and Laboratory Standard Institutes (CLSI-2016) criteria. E. coli ATCC 25922 was used as the control for antimicrobial susceptibility testing.
Detection of resistance genes
DNA was extracted from the bacteria using the boiling method. Polymerase chain reaction (PCR) and nucleotide sequencing techniques were performed in 10 CREL strains to confirm the existence of carbapenemase genes blaKPC, blaNDM, blaVIM-1, blaVIM-2, blaIMP-4, blaIMP-8, blaOXA-1, blaOXA- 23, blaOXA-24, blaOXA-48, blaOXA-51, blaOXA-58, and ESBL genes, including blaCTX, blaTEM, blaACC, and blaSHV, using primers as previously described and listed at Table 1 (6). According to the size of the target fragment, positive amplification products were sequenced, and the sequencing results were compared against the BLAST tool.

Full table
Multilocus sequence typing (MLST)
MLST was performed using seven housekeeping genes of Enterobacter cloacae, which were amplified using primers shown in online databases (https://pubmlst.org/ecloacae/). The products of PCR were sequenced. Sequence types (STs) were determined using online database tools. Molecular Phylogenetic analysis by Maximum Likelihood method, evolutionary analyses were conducted in MEGA X software.
Statistical analysis
The SPSS 25.0 software was conducted using the Chi-square test for statistical analysis. P<0.05 was statistically significant.
Results
Clinical data
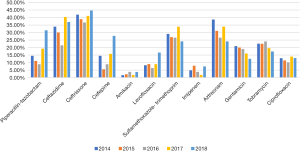
From January 2014 to December 2018, a total of 342 non-repetitive Enterobacter cloacae were detected. The most common specimens were sputum (202/342, 59%), followed by secretion (41/342, 12%), blood (39/342, 11.4%). The most common clinical departments were respiratory, followed by neurosurgery, emergency ICU, orthopedics, and emergency department (Table 2). CREL information was shown in Tables 3 and 4. The trends of resistance rate of Enterobacter cloacae were shown in Figure 1.

Full table
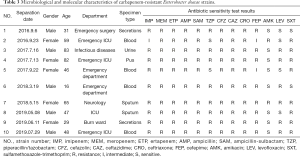
Full table
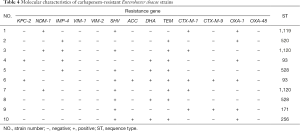
Full table
Antimicrobial susceptibility test
As shown in Table 5, amikacin was the lowest in the five years, and the resistance rate of Enterobacter cloacae to ceftriaxone was the highest. The resistance rate of the fourth-generation cephalosporin cefepime has gradually increased since 2015. Both Piperacillin/Tazobactam and Cefepime have statistically difference in the resistance rates in the five years. Moreover, compared with non-CREL, the resistance rate of CREL strains was significantly higher (P<0.05)
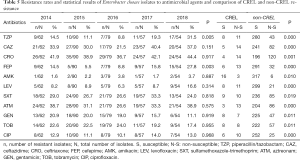
Full table
Molecular analyses
Among the ten strains, three types of carbapenemases were confirmed, including IMP-4, KPC-2, NDM-1. The corresponding numbers of the strains that produced the primary types of carbapenemases were 4, 3, 2. Other carbapenemase genes were not detected, with one strain co-producing NDM-1 and IMP-4. TEM and SHV were the most often ESBL gene, followed by OXA-1.
MLST
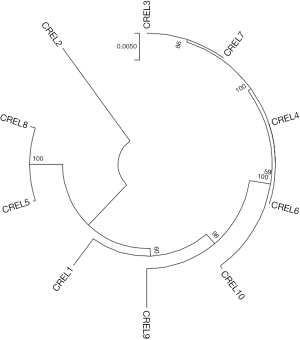
The results of the MLST are shown in Table 3. A total of 7 sequence types were detected in the 10 CREL strains. ST93 (2/10), ST528 (2/10), and ST1120 (2/10) were isolated two of each ST, while the others were assigned to an isolate per ST. The phylogenetic tree built from the sequence data of the seven MLST genes showed that ST93, ST1120, ST256 has high homology (ST256 and ST1120 differed from ST93 by one and two housekeeping genes, respectively). The phylogenetic tree, as shown in Figure 2.
Discussion
Enterobacter cloacae is an important opportunistic pathogen and frequently implicated in nosocomial infections (7). The carrying of drug-resistant genes also leads to the multidrug-resistance and limiting therapeutic options even further (8). Our study showed that most of the isolates remained susceptible to tested antibiotics; however, CREL holds a significantly higher resistance rate compared with non-CREL. The drug resistance rate of Cefepime has been increasing in recent years, from 5.5% in 2015 to 27.8% in 2018. Previous research has reported that the combination of different resistance mechanisms, such as porin loss (primarily OmpC), expression of blaOXA-1, and/or TEM-1, could confer decreased susceptibility to Cefepime (9-11). Cefepime resistance has been described in Pseudomonas aeruginosa and E.col strains producing OXA-1 beta-lactamase (12,13). In our study, 6 of 10 strains carrying the blaOXA-1 gene, which may be one of the reasons for the increased resistance rate of Cefepime in recent years. However, we only investigated CREL strains, the carrying rate of the blaOXA-1 gene in non-CREL strains, and other mechanisms that need further exploration.
Carbapenemase spread has been increasingly reported worldwide over the last decade (14). VIM-1 was most common in Spain and other southern European countries (15), in the current study, no producers of VIM-1 were found. A previous multicenter study conducted a molecular epidemiological survey of carbapenem-resistant Enterobacter cloacae (CREL) in 11 cities in China to understand the prevalence of the bacteria further. E. cloacae prevalent in China produced NDM-1 and IMP-4. NDM-1 was found in the highest proportion and may represent a significant drug-resistant mechanism of carbapenem-producing Enterobacteriaceae in China (1). In the present study, a higher proportion (4/10, 40%) of blaIMP-4 positive strains were identified among carbapenem-resistant E. cloacae isolates showing that IMP-4 was the dominant carbapenemase instead of NDM-1 in this region. Australia has observed the dominance of IMP-4-producing Enterobacter cloacae, which agreed with the current study (16). It is worth noting that one strain of this study co-producing NDM-1 and IMP-4. One CREL strain was previously reported to co-producing NDM-1 and IMP-8 in Chongqing, China (17). NDM and IMP types belong to class B metallo-lactamases (MBLs) in the Ambler classification and disseminated among bacteria internationally (18). Considering that one strain co-producing NDM-1 and IMP-4 may confer multiple resistance to antibiotics that further reduce the therapeutic choices, surveillance for carbapenemase detection, and infection control measures should be implemented to prevent their further spread. The OXA-48 carbapenemase was first discovered in various Enterobacteriaceae species isolated in Turkey and other countries in the Middle East, also be reported in Enterobacter cloacae (19). It belongs to the D-class carbapenemase of Ambler classification, which has a weak role in the hydrolysis of carbapenems. OXA-48 was not detected in our study, showing that it is not prevalent in our region. TEM and SHV was the dominant ESBL type in our study, unlike CTX-M-producers most frequent in Latin American countries (20).
MLST showed subtype diversity. Our study has revealed that local carbapenem-resistant E. cloacae isolates did not evolve from a unique ancestral background. A total of seven sequence types were detected in 10 carbapenem-resistant E. cloacae strains. Previous studies have reported some sporadic cases of E. cloacae isolates harboring NDM-1, such as ST92 in Croatia, ST265 in Australia (21,22). Our study identified a potential prevalent clone of ST1120 carbapenem-resistant E. cloacae isolates carrying NDM-1. However, this ST was different from some widespread E. cloacae STs (ST66, ST78, ST108, and ST114) that reported in European countries (23). The primary epidemic strains in Shenzhen city and Henan province in china were ST418 and ST120, respectively (1,24). The outbreak of ST88 carbapenem-resistant E. cloacae isolates carrying NDM-1 was reported in Chongqing, China (17). The first IMP-4-Producing Enterobacter cloacae sequence type 74 and 194 have reported in Korea in 2017 (25). Our results showed that four strains of IMP-4-producing assigned different ST typing revealed the genetic diversity of carbapenem-resistant E. cloacae. KPC carbapenemase is primarily found in Klebsiella pneumoniae, while KPC-producing E. cloacae infections have been relatively infrequent (26). However, the outbreak of KPC-producing E. cloacae ST114 has been reported in the United States (27). In China, the first KPC-producing CREL appeared in Shanghai in 2010 (28). In the present study, two KPC-producing isolates are assigned to ST93. Moreover, the MEGA analysis showed that ST93, ST256, and ST1120 have high homology, showing that CREL in our area has a potential spread risk. Since limited numbers were collected, the NDM-1-possessing ST1120 isolates, KPC-producing ST93 strains, and IMP-4-producing strains in our region should be taken with serious concern and still need to be further monitored.
In conclusion, our study investigated antibiotic susceptibility and molecular analyses of clinical Enterobacter cloacae isolated in Eastern Heilongjiang Province, China. The results showed that antimicrobial resistance was low among them. Compared with non-CREL, the resistance rate of CREL strains was significantly higher. ST93, ST1120, ST256 has high homology showing that CREL in our area has potential spread risk. The NDM-1-possessing ST1120 isolates, KPC-producing ST93 strains, and IMP-4-producing strains in our region should be taken with serious concern s and still need to be further monitored.
There are some limitations in this study: (I) we did not determine the exact mechanism of how each E. cloacae acquired the carbapenemase. Further studies are necessary to determine the exact mechanisms by which E. cloacae acquired the carbapenemase. (II) The number of CREL collected was limited, and we only investigated one hospital, so our conclusion may not be comprehensive enough or extended directly to the whole region.
Acknowledgments
Funding: This work was supported by Excellent Team of Young Teachers Foundation of Heilongjiang Province (2018-KYYWF-0916), Research Project of Yongchuan Hospital Affiliated to Chongqing Medical University (YJYJ201902).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-1089
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-1089). The authors report grants from Excellent Team of Young Teachers Foundation of Heilongjiang Province, grants from Research Project of Yongchuan Hospital Affiliated to Chongqing Medical University, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhao Y, Li C, Zhang J, et al. The in vitro activity of polymyxin B and tigecycline alone and combination with other antibiotics against carbapenem-resistant Enterobacter cloacae complex isolates, including high-risk clones. Ann Transl Med 2019;7:779. [Crossref] [PubMed]
- Babouee Flury B, Ellington MJ, Hopkins KL, et al. Association of Novel Nonsynonymous Single Nucleotide Polymorphisms in ampD with Cephalosporin Resistance and Phylogenetic Variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae Isolates That Are Highly Resistant to Carbapenems. Antimicrob Agents Chemother 2016;60:2383-90. [Crossref] [PubMed]
- van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017;8:460-9. [Crossref] [PubMed]
- Potter RF, D'Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat 2016;29:30-46. [Crossref] [PubMed]
- Hayakawa K, Miyoshi-Akiyama T, Kirikae T, et al. Molecular and epidemiological characterization of IMP-type metallo-beta-lactamase-producing Enterobacter cloacae in a Large tertiary care hospital in Japan. Antimicrob Agents Chemother 2014;58:3441-50. [Crossref] [PubMed]
- Gong X, Zhang J, Su S, et al. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect Dis 2018;18:417. [Crossref] [PubMed]
- Wang S, Xiao SZ, Gu FF, et al. Antimicrobial susceptibility and molecular epidemiology of clinical Enterobacter cloacae bloodstream isolates in Shanghai, China. PLoS One 2017;12:e0189713. [Crossref] [PubMed]
- Lahlaoui H, Anis BH, Mohamed K, et al. Emergence of SHV-12 extended spectrum beta-lactamase among clinical isolates of Enterobacter cloacae in Tunisia. Microb Pathog 2012;53:64-5. [Crossref] [PubMed]
- Torres E, Lopez-Cerero L, Rodriguez-Martinez JM, et al. Reduced Susceptibility to Cefepime in Clinical Isolates of Enterobacteriaceae Producing OXA-1 Beta-Lactamase. Microb Drug Resist 2016;22:141-6. [Crossref] [PubMed]
- Yasufuku T, Shigemura K, Shirakawa T, et al. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli Strains clinically isolated from urinary tract infection patients. J Clin Microbiol 2011;49:189-94. [Crossref] [PubMed]
- Liu YF, Yan JJ, Lei HY, et al. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect Immun 2012;80:1815-22. [Crossref] [PubMed]
- Aubert D, Poirel L, Chevalier J, et al. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2001;45:1615-20. [Crossref] [PubMed]
- Ortega A, Oteo J, Aranzamendi-Zaldumbide M, et al. Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 2012;56:3576-81. [Crossref] [PubMed]
- Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol 2007;2:501-12. [Crossref] [PubMed]
- Villa J, Viedma E, Branas P, et al. Multiclonal spread of VIM-1-producing Enterobacter cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int J Antimicrob Agents 2014;43:451-5. [Crossref] [PubMed]
- Sidjabat HE, Townell N, Nimmo GR, et al. Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother 2015;59:4059-66. [Crossref] [PubMed]
- Jia X, Dai W, Ma W, et al. Carbapenem-Resistant E. cloacae in Southwest China: Molecular Analysis of Resistance and Risk Factors for Infections Caused by NDM-1-Producers. Front Microbiol 2018;9:658. [Crossref] [PubMed]
- Tzouvelekis LS, Markogiannakis A, Psichogiou M, et al. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 2012;25:682-707. [Crossref] [PubMed]
- Adler A, Solter E, Masarwa S, et al. Epidemiological and microbiological characteristics of an outbreak caused by OXA-48-producing Enterobacteriaceae in a neonatal intensive care unit in Jerusalem, Israel. J Clin Microbiol 2013;51:2926-30. [Crossref] [PubMed]
- Seki LM, Pereira PS, de Souza Conceicao M, et al. Molecular epidemiology of CTX-M producing Enterobacteriaceae isolated from bloodstream infections in Rio de Janeiro, Brazil: emergence of CTX-M-15. Braz J Infect Dis 2013;17:640-6. [Crossref] [PubMed]
- Zujić Atalić V, Bedenic B, Kocsis E, et al. Diversity of carbapenemases in clinical isolates of Enterobacteriaceae in Croatia--the results of a multicentre study. Clin Microbiol Infect 2014;20:O894-903. [Crossref] [PubMed]
- Wailan AM, Paterson DL, Kennedy K, et al. Genomic Characteristics of NDM-Producing Enterobacteriaceae Isolates in Australia and Their blaNDM Genetic Contexts. Antimicrob Agents Chemother 2015;60:136-41. [Crossref] [PubMed]
- Izdebski R, Baraniak A, Herda M, et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 2015;70:48-56. [Crossref] [PubMed]
- Liu C, Qin S, Xu H, et al. New Delhi Metallo-beta-Lactamase 1(NDM-1), the Dominant Carbapenemase Detected in Carbapenem-Resistant Enterobacter cloacae from Henan Province, China. PLoS One 2015;10:e0135044. [Crossref] [PubMed]
- Lee JH, Bae IK, Lee CH, et al. Molecular Characteristics of First IMP-4-Producing Enterobacter cloacae Sequence Type 74 and 194 in Korea. Front Microbiol 2017;8:2343. [Crossref] [PubMed]
- Centers for Disease C. Prevention. Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep 2013;62:165-70. [PubMed]
- Kanamori H, Parobek CM, Juliano JJ, et al. A Prolonged Outbreak of KPC-3-Producing Enterobacter cloacae and Klebsiella pneumoniae Driven by Multiple Mechanisms of Resistance Transmission at a Large Academic Burn Center. Antimicrob Agents Chemother 2017;61:e01516-16. [Crossref] [PubMed]
- Wu Q, Liu Q, Han L, et al. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn Microbiol Infect Dis 2010;66:326-8. [Crossref] [PubMed]