
Characterization of a multidrug-resistant Klebsiella pneumoniae ST3330 clone responsible for a nosocomial outbreak in a neonatal intensive care unit
Introduction
Neonatal septicemia, which is mainly hospital-acquired, especially in neonatal intensive care units (NICUs), is becoming more challenging for practitioners to control (1,2). Septicemia and pneumonia are the main symptoms of neonatal sepsis (3).
Klebsiella pneumoniae (Kp) is a common cause of blood infections. Multidrug-resistant (MDR) strains of Kp, notably extended-spectrum-beta-lactamase (ESBL)-producing strains, are regularly involved in nosocomial outbreaks (3-7). These strains are able to acquire additional resistance mechanisms and virulence factors via plasmids in particular, which enables them to disseminate across healthcare units and increase their invasive power in the human body (5,8). Transmission can occur either from the mother to child at birth, or acquired during nursery by person-to-person transmission, via the hands of the nursing staff and the contaminated equipment, food or the environment. Rigorous nosocomial infection prevention measures can be developed to control klebsiella pneumoniae in NICU. Carbapenems are the last antibiotics to control klebsiella pneumoniae infection. Treatment of klebsiella pneumoniae infection in children can draw on the experience of treatment in adults, but drug dosage needs to be adjusted. In neonates, ESBL-KP infections, which carry high morbidity and mortality (3), have become a burden and a threat to public health (5,9).
It is crucial that the source of emerging multi-drug bacteria is identified. Therefore, it is vital that strain-level discrimination of the involved pathogens is carried out and the bacteria at the core of any outbreak is characterized (3). This study aimed to conduct an examination of the phenotypes, genotypes, and genetic relatedness of ESBL-KP that caused an outbreak of sepsis among neonates in an intensive care unit of a Beijing hospital. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/apm-20-958).
Methods
Patients’ clinical and epidemiological features
Between April 2016 and May 2018, 21 strains (numbered 1–21) were collected from 21 newborns aged 1–33 days who were hospitalized in the NICU in a Beijing Hospital (Table 1). All 21 strains were isolated from the bloodstreams of different patients. A descriptive study based on electronic records was conducted to describe demographic data and clinical events related to ESBL-Kp infections (1,5). As the clinical samples were routinely taken for bacterial testing purposes in public hospitals, and the data were analyzed anonymously, ethical approval was not required (3).

Full table
Phenotypic characterization
Blood cultures were monitored with the Bactec Microbial Detection System (Bactec 9050, Becton-Dickinson Company, 1 Becton Drive, Franklin Lakes, New Jersey). Bacterial identification was performed by MALDI-ToF mass spectrometry (MALDI Biotyper; Bruker Daltonics, France). An automated VITEK 2 compact system (BioMérieux, Inc.) was employed to test antibiotic susceptibility. The measurement of MICs of antimicrobial agents of common clinical usage was conducted using a VITEK 2 AST-GN09 test Kit (BioMérieux, Inc.) in line with the protocol of the manufacturer. The interpretation of the results of all susceptibility testing was guided by the Clinical and Laboratory Standards Institute (CLSI) guidelines (3,10,11). In antimicrobial susceptibility testing, we chose E. coli ATCC 25922 as the control strain (12). Phenotypic ESBL detection was performed according to the combined antibiotic disks method (between third-generation cephalosporins and clavulanic acid) and by observing ceftriaxone susceptibility (5). Patient demographics and susceptibility data were analyzed with WHONET software (version 5.6).
Molecular detection of resistance genes
Polymerase chain reaction (PCR) assays were performed to detect Drug resistance genes, including carbapenemases (blaKPC, blaAIM, blaGIM, blaSIM, blaNDM, blaIMP, blaVIM, and blaOXA-48), common extended-spectrum-beta-lactamase (ESBL) genes (blaCTX-M, blaTEM, blaOXA, and blaSHV). The PCR primers previously described (5,11-13) were listed in Table 2. The cycling parameters were as follows: 94 °C for 10 min followed by 36 cycles of 94 °C for 30 s, 52 °C for 40 s and 72 °C for 50 s, with a final extension at 72 °C for 5 min. Electrophoresis with a 1.5% agarose gel was carried out to screen the positive amplicons, which were then sequenced. Analysis and comparisons were then made between the DNA sequences obtained and those available on the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) via BLAST searches (13).
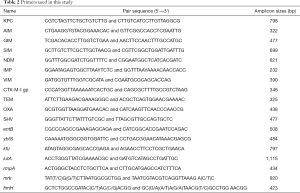
Full table
Detection of virulence-associated genes
Genes encoding specific virulence factors involved in iron acquisition systems (entB, ybtS, kfu, and iutA), hyper-mucoviscous phenotype (rmpA), and fimbrial adhesions (mrk and fimH) were researched using a multiplex PCR, as previously described (5,14,15). The PCR primers were listed in Table 2. The cycling parameters were as follows: 94 °C for 5 min followed by 35 cycles of 94 °C for 1 min, 50 °C for 30 s and 72 °C for 50 s, with a final extension at 72 °C for 5 min. The results were analyzed by BLAST program.
Pulsed-field gel electrophoresis (PFGE) analyses
PFGE with XbaI was conducted for each clinical Kp isolate, as previously described (11,12,16,17). A lambda ladder pulsed-field gel marker was used as a size standard for PFGE. The identification and interpretation of the results were conducted according to the Tenover criteria. Isolates with genetic similarity of ≥95% were judged to be the same strain (PFGE type) (3).
Multilocus sequence typing (MLST)
Multilocus sequence typing was performed by PCR amplifying and sequencing seven housekeeping genes according to protocols provided on the MLST website for K. pneumoniae (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) (11,16,17). Sequences of seven housekeeping genes were obtained for isolates from all PFGE types. Alleles and STs were assigned accordingly. Sequences of any alleles that were not on the database were submitted to the curator and new allele numbers were obtained. Strains with a difference in two or more alleles were considered to be unrelated. A DNA analyzer (3730xl; Applied Biosystems, Life Technologies, Foster City, CA, USA) was employed to sequence each amplification product (12).
Biofilm formation assay
The standard static biofilm assay described by Naparstek et al. (18) was performed on the isolates. Briefly, the isolates underwent incubation at 37 °C for 18 h, after which 200 µL of broth was transferred to each well of a sterile phenotypic microplate (96 cells). Next, the addition of 10 µL of bacterial suspension from the incubated isolates at an opacity 0.5 McFarland was made to the wells. The microplate was incubated at 37 °C for 24 h and washed with saline 3 times to keep the plate completely dry. After that, 200 µL of 0.1% crystal violet was added to the wells for 20 min and the microplate was washed 3 times with saline and dried. Finally, the addition of 200 µL of 95% ethanol was made to each well, and the plate was read with ELISA Plate Reader at 590 nm. Biofilm formation is indicated by high absorption (18-20). All experiments were repeated in triplicate. A well without bacterial suspension served as a negative control.
Serum killing assay
The blood samples of 10 healthy adults were collected for the serum killing assay. An inoculum of 25 µL (adjusted to 106 colony forming units/mL) prepared from the mid-log phase was diluted by 0.9% saline and added into a 10×75 mm Falcon polypropylene tube (BD Biosciences, Franklin Lakes, New Jersey) containing 75 µL of pooled human sera. The tubes underwent agitation for 0, 60, 120, or 180 min. After being exposed to serum, the number of viable bacteria were determined by removing an aliquot of each bacterial suspension at the designated time point, adding broth to dilute it 10-fold, plating it on Mueller-Hinton agar, and performing the assay, as described immediately below. Each strain was tested at least 3 times (14). The results were expressed as percentage of inoculation and the responses in relation to viable counts were graded from 1 to 6, as previously described. All isolates were classified as follows: highly sensitive (grades 1 or 2), intermediately sensitive (grades 3 or 4), or resistant (grades 5 or 6) (21).
Statistical analysis
Biofilm formation in the ST3330 group was compared with that in the other STs group, and differences were analyzed by unpaired t-test (18). A P value <0.05 represented statistical significance. All statistical analyses were carried out with GraphPad Prism, version 8.0 (GraphPad Software, San Diego, CA, USA).
Results
Clinical and epidemiological features
The epidemic period extended from April 2016 to May 2018. Twenty-one patients were positive for ESBL-Kp colonization and infection. One patient died of severe infection by clinical diagnosis. All patients were hospitalized on the same floor and supported by the same staff. During the study period a total of 814 neonates were admitted to the NICU including 426 (52.3%) who were isolated with gram-negative pathogens. K. pneumoniae accounted for 47% of these pathogens. The twenty-one patients clinical data are shown in Table 1.
Antimicrobial susceptibility testing and detection of resistance mechanisms
All strains had an ESBL phenotype and were therefore resistant to all third-generation cephalosporins and fully susceptible to carbapenems (Table 3). Among the 21 ESBL-Kp isolates, 16 produced β-lactamases (CTX-M-3 and TEM-1B), while the others possessed CTX-M-15, CTX-M-24, CTX-M-66, TEM-1C, SHV-26, SHV-172, and OXA-1 (Table 3).
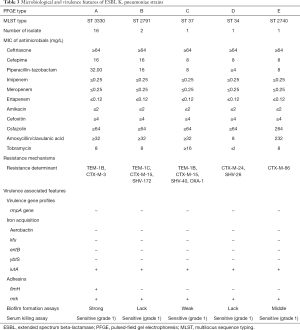
Full table
PFGE and MLST
Pulsed-field gel electrophoresis (PFGE) analysis of the 21 ESBL-KP isolates showed 21 banding patterns classified into 5 PFGE types (A, B, C, D and E) as shown in Figures 1 and 2, which is confirmed by MLST (Table 3).
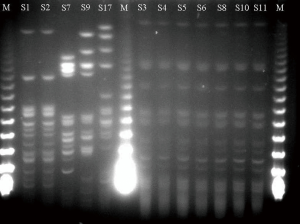
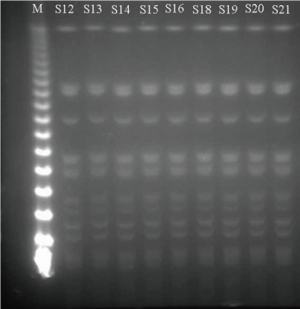
Detection of virulence-associated determinants, biofilm formation, and serum killing
Despite the reported association between the rmpA gene and hypermucoviscosity, which has always been associated with more virulent strains, the rmpA gene was not detected in any of the isolates. All the 21 strains tested negative for aerobactin, suggesting low potential for hypervirulence. Our analysis also took other virulence gene profiles into account. The virulence genes iutA and mrk were detected in all of the isolates. PFGE type A carried both mrk (type 3 fimbriae, biofilm formation) and fimH (type 1 fimbriae), which indicated stronger biofilm formation. A summary of the virulence-associated genes is given in Table 3.
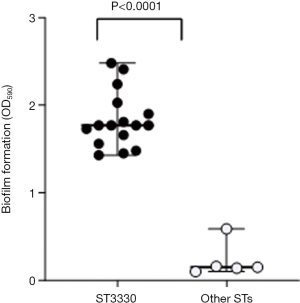
Biofilm formation assays were performed on all 21 ESBL-Kp isolates and revealed relatively high-mass biofilms (OD590 range, 0.0994–2.4756). It was discovered that isolates with the endemic ST3330 lineage produced more biofilm in comparison to other ST isolates (median OD590 1.829 vs. 0.2280, respectively; P<0.0001) (Figure 3).
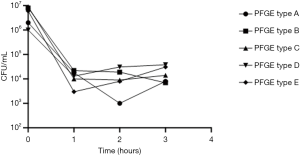
Serum killing assay found that five PFGE types of isolates showed serum high sensitivity (grade 1) (Figure 4).
Discussion
Premature newborns hospitalized in intensive care units are in a vulnerable and immunocompromised state. Moreover, intensive nursing is an additional risk factor for nosocomial infections (22,23). This study collected and analyzed original data from the neonatal unit at one Beijing hospital, and attests to the commonness of neonatal sepsis, as well as its association with high risk of mortality. In neonatal sepsis, the predominance of gram-negative pathogens (47% of strains were K. pneumonia in our hospital) indicates that infections are most likely horizontally transmitted from the environment and healthcare providers (1,5-7,24). Establishing whether a positive growth in the blood culture is a pathogen or a contaminant is paramount and a number of parameters need to be taken into account before a conclusion is reached. To date, the increased emergence of MDR organisms has limited treatment choices and affected their timeliness and efficiency. Therefore, because of the lack of suitable antibiotics for pediatric patients, the treatment of these infections presents an ever-increasing challenge for pediatricians (4,25,26).
In this investigation, PFGE type A surfaced as the most prevalent strain that disseminated within two years. The type A strain was revealed by MLST analysis to be ST3330 (Table 3). Therefore, there was evidence to suggest that the ESBL-K.pneumoniae isolates in this study had clonal dissemination. Moreover, an association was identified between ST3330 and the spread and outbreak of CTX-M-3 and TEM-1-producing K. pneumoniae. This finding is at odds with a previous report that demonstrated CTX-M-15 to be the most prevalent ESBL gene in the world (5,27,28). CTX-M-3 and CTX-M-15 belong to the CTX-M 1 group. CTX-M-15 derives from the mutation of CTX-M-3 at the 240th amino acid from aspartic acid to glycine. An earlier Italian study demonstrated the existence of Kp strain co-harboring genes blaTEM-1 and blaCTX-M-15 (5). Although type B and type C produced CTX-M-15, SHV, and OXA, they were not prevalent genes in this study. Generally, CTX-M and TEM confer resistance to third-generation cephalosporins but cannot reduce their susceptibility to carbapenems (5,9). Most of the clinical strains in the current study demonstrated high-level resistance to all third-generation cephalosporins and full susceptibility to carbapenems (Table 3).
Several Kp clones, such as ST23, are described as hypervirulent (hvKP) and regularly show a hypermucoviscous phenotype on agar plates. These hypermucoviscous strains are linked to production of several specific capsular serotypes (mainly K1 and K2) and the “regulator of mucoid phenotype A” (rmpA/rmpA2) complex (10,29). ESBL production is understood to have an association with a greater expression of virulence factors, including cell invasion proteins and fimbrial adhesins (30). Moreover, various elements may render the strain more virulent, such as biofilm formation (with fimbriae), additional iron acquisition systems (siderophores), or allantoin metabolism. The mrkABCDF gene cluster in our strains and the presence of the iutA gene that encode adhesin type 3 fimbriae and bacterial siderophores, respectively, bear a strong association with biofilm formation (30). Most of the ST3330 strains involved in this dissemination carried at least two fimbriae: mrk (type 3 fimbriae) and fimH (type 1 fimbriae). Fimbriae are adhesive structures on the surface of Kp involved in biofilm formation. Notably, type 1 (encoded by the fim operon) and type 3 fimbriae (encoded by the mrk operon) are considered to be virulence factors expressed by almost all Kp strains (5,18). Biofilm formation assays performed on all 21 ESBL-Kp isolates showed relatively high-mass biofilms. Isolates with endemic ST3330 lineage were found to produce a larger amount of biofilm than other ST isolates (P<0.0001) (Figure 3). A pediatric patient was assumed to be the source of the outbreak, but the pathogen still was being disseminated after the patient’s discharge. Furthermore, the Hospital Infection Control Department did not isolate the epidemic strain during two years of environmental sampling. Strong biofilm formation ability may be a cause of unsuccessful sampling. It may partly explain the epidemic strain has high colonization and diffusion potential. The source of the infection is still unclear and needs to be traced through future prospective study.
In this study, each of the 21 strains was negative for rmpA and aerobactin, which indicates low potential for hypervirulence. Meanwhile, serum killing assay showed five PFGE types of isolates to present serum high sensitivity (grade 1) (Figure 4) (14,21). As it was impossible to collect serum from healthy newborns, human serum was obtained from 10 healthy adults, which may have led to errors. The first-line empirical treatment is Cefoperazone Sodium and Sulbactam Sodium combined with third-generation cephalosporin. In the absence of any improvement, antibiotics, carbapenems, and vancomycin are used until the targeted blood culture results are obtained. In general, the majority outcomes of these newborns were consistent with clinical treatment excluding the case of one newborn who died of severe infection by clinical diagnosis. However, even in this case, it was difficult to establish if the infection was the only cause of death.
The present study has several limitations. First, its single-center nature and relatively small sample size mean that it might lack either the scientific precision or external validity needed to bring about widespread changes in clinical practice. Second, the samples were collected from a single hospital in Beijing. Therefore, future work including samples from a larger geographic area or a multi-center analysis is suggested.
Conclusions
Neonatal sepsis was observed and associated with elevated inpatient mortality. In neonates, sepsis can be caused by several microorganisms, among which, K. pneumonia is proved to be the most frequently detected pathogen. K. pneumonia has the capacity to develop multi-antibiotic resistance, including to third generation cephalosporins. Because of the gaps in disease records, more data on the microbiology of neonatal infection need to be collected to bring about improvements to existing practices. Hospitals should adopt approaches aimed at preventing and managing the spread of infection. Emergent ESBL-Kp infections can be traced at the earliest stage, by employing infection-control measures, such programs for stewarding antibiotics with continuous surveillance, especially in at-risk units such as NICUs.
Acknowledgments
Thanks to Bei Yao at the Department of Clinical Laboratory, Peking University Third Hospital for collecting samples, and to Jiyong Yang at Department of Clinical Laboratory, Medical Laboratory Center, Chinese PLA General Hospital for proof reading the article.
Funding: This work was supported by the National Natural Science Foundation of China (No. 81871731).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-958
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-958
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-958). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was not required.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gao K, Fu J, Guan X, et al. Incidence, Bacterial Profiles, And Antimicrobial Resistance Of Culture-Proven Neonatal Sepsis In South China. Infect Drug Resist 2019;12:3797-805. [Crossref] [PubMed]
- Yin D, Zhang L, Wang A, et al. Clinical and molecular epidemiologic characteristics of carbapenem-resistant Klebsiella pneumoniae infection/colonization among neonates in China. J Hosp Infect 2018;100:21-8. [Crossref] [PubMed]
- Ghaith DM, Zafer MM, Said HM, et al. Genetic diversity of carbapenem-resistant Klebsiella Pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur J Clin Microbiol Infect Dis 2020;39:583-91. [PubMed]
- Bassetti M, Giacobbe DR. Reducing dissemination of carbapenem-resistant Klebsiella pneumoniae. Ann Transl Med 2019;7:S365. [Crossref] [PubMed]
- Peltier F, Choquet M, Decroix V, et al. Characterization of a multidrug-resistant Klebsiella pneumoniae ST607-K25 clone responsible for a nosocomial outbreak in a neonatal intensive care unit. J Med Microbiol 2019;68:67-76. [Crossref] [PubMed]
- Patil S, Chen H, Zhang X, et al. Antimicrobial Resistance and Resistance Determinant Insights into Multi-Drug Resistant Gram-Negative Bacteria Isolates from Paediatric Patients in China. Infect Drug Resist 2019;12:3625-34. [Crossref] [PubMed]
- Oliveira PMN, Buonora SN, Souza CLP, et al. Surveillance of multidrug-resistant bacteria in pediatric and neonatal intensive care units in Rio de Janeiro State, Brazil. Rev Soc Bras Med Trop 2019;52:e20190205. [Crossref] [PubMed]
- Leikin-Zach V, Shany E, Yitshak-Sade M, et al. Neonatal Risk Factors for Colonization with Extended-Spectrum Beta-Lactamase-Producing Bacteria in the Neonatal Intensive Care Unit. Isr Med Assoc J 2018;20:286-90. [PubMed]
- Uc-Cachón AH, Gracida-Osorno C, Luna-Chi IG, et al. High Prevalence of Antimicrobial Resistance Among Gram-Negative Isolated Bacilli in Intensive Care Units at a Tertiary-Care Hospital in Yucatan Mexico. Medicina (Kaunas) 2019;55:588. [Crossref] [PubMed]
- An J, Lai K, Ma Y, et al. Emergence of multiple carbapenemase-producing organisms in single patients: an increasing threat to treatment of infection. J Antimicrob Chemother 2018;73:544-6. [Crossref] [PubMed]
- Tian D, Pan F, Wang C, et al. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist 2018;11:1935-43. [Crossref] [PubMed]
- Lai K, Ma Y, Guo L, et al. Molecular characterization of clinical IMP-producing Klebsiella pneumoniae isolates from a Chinese Tertiary Hospital. Ann Clin Microbiol Antimicrob 2017;16:42. [Crossref] [PubMed]
- Ma Y, Bao C, Liu J, et al. Microbiological characterisation of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Beijing, China. J Glob Antimicrob Resist 2018;12:162-6. [Crossref] [PubMed]
- Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015;71:553-60. [Crossref] [PubMed]
- Luo Y, Ma Y, Zhao Q, et al. Similarity and divergence of phylogenies, antimicrobial susceptibilities, and virulence factor profiles of Escherichia coli isolates causing recurrent urinary tract infections that persist or result from reinfection. J Clin Microbiol 2012;50:4002-7. [Crossref] [PubMed]
- Yang J, Ye L, Guo L, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 2013;19:E509-15. [Crossref] [PubMed]
- Guo L, An J, Ma Y, et al. Nosocomial Outbreak of OXA-48-Producing Klebsiella pneumoniae in a Chinese Hospital: Clonal Transmission of ST147 and ST383. PLoS One 2016;11:e0160754. [Crossref] [PubMed]
- Naparstek L, Carmeli Y, Navon-Venezia S, et al. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 2014;69:1027-34. [Crossref] [PubMed]
- Huertas MG, Zarate L, Acosta IC, et al. Klebsiella pneumoniae yfiRNB operon affects biofilm formation, polysaccharide production and drug susceptibility. Microbiology 2014;160:2595-606. [Crossref] [PubMed]
- Khodadadian R, Rahdar HA, Javadi A, et al. Detection of VIM-1 and IMP-1 genes in Klebsiella pneumoniae and relationship with biofilm formation. Microb Pathog 2018;115:25-30. [Crossref] [PubMed]
- Abate G, Koh TH, Gardner M, et al. Clinical and bacteriological characteristics of Klebsiella pneumoniae causing liver abscess with less frequently observed multi-locus sequences type, ST163, from Singapore and Missouri, US. J Microbiol Immunol Infect 2012;45:31-6. [Crossref] [PubMed]
- Arena F, Giani T, Becucci E, et al. Large oligoclonal outbreak due to Klebsiella pneumoniae ST14 and ST26 producing the FOX-7 AmpC beta-lactamase in a neonatal intensive care unit. J Clin Microbiol 2013;51:4067-72. [Crossref] [PubMed]
- Cadot L, Bruguiere H, Jumas-Bilak E, et al. Extended spectrum beta-lactamase-producing Klebsiella pneumoniae outbreak reveals incubators as pathogen reservoir in neonatal care center. Eur J Pediatr 2019;178:505-13. [Crossref] [PubMed]
- Devrim F, Serdaroglu E, Caglar I, et al. The Emerging Resistance in Nosocomial Urinary Tract Infections: From the Pediatrics Perspective. Mediterr J Hematol Infect Dis 2018;10:e2018055. [Crossref] [PubMed]
- Zou ZH, Liu D, Li HD, et al. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann Clin Microbiol Antimicrob 2018;17:9. [Crossref] [PubMed]
- Corbella M, Caltagirone M, Gaiarsa S, et al. Characterization of an Outbreak of Extended-Spectrum beta-Lactamase-Producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in Italy. Microb Drug Resist 2018;24:1128-36. [Crossref] [PubMed]
- Weissman SJ, Adler A, Qin X, et al. Emergence of extended-spectrum beta-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int J Antimicrob Agents 2013;41:414-20. [Crossref] [PubMed]
- Justo-da-Silva LH, De-Azeredo AN, Bueno AC, et al. Diversity of clonal types of Klebsiella pneumoniae causing infections in intensive care neonatal patients in a large urban setting. Braz J Microbiol 2019;50:935-42. [Crossref] [PubMed]
- Lin JC, Koh TH, Lee N, et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog 2014;6:21. [Crossref] [PubMed]
- Nordberg V, Jonsson K, Giske CG, et al. Neonatal intestinal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae-a 5-year follow-up study. Clin Microbiol Infect 2018;24:1004-9. [Crossref] [PubMed]


