Genetic biomarkers associated with changes in quality of life and pain following palliative radiotherapy in patients with bone metastases
Introduction
Bone is one of the most common sites with metastases, and many cancer patients with metastases to the bone can live for years upon their initial diagnosis (1). However, the development of metastatic loci in bone can lead to pain and fracture risk, which greatly impede quality of life (QOL) (2). QOL encompasses multiple functional and symptomatic domains of life such as nausea and vomiting, fatigue, and pain (3). In particular, the presence of pain can negatively affect almost all other aspects of QOL, including the functional and emotional domains (4). Since patients with bone metastases often have survival measured in years, maintaining their QOL through managing symptoms such as pain that are associated with disease and treatment is of increasing clinical importance.
Radiotherapy is effective in the palliation of bone metastases to improve QOL and manage pain. However, there is variability in patient response. Studies have found that altered protein levels, particularly of inflammatory proteins, are associated with variations in QOL, therefore pointing to genetic variation as a potential underlying cause that explains inter-individual differences (5). Moreover, family and twin studies suggest that around 30–50% of patient-reported QOL aspects is heritable, strengthening the argument for genetic predisposition of QOL (6,7).
Single nucleotide variants are differences at individual nucleic acid bases in DNA in the population, and represent the most common form of genetic variation (8). Therefore, it has been the object of association studies which identify genetic biomarkers predictive of QOL. For example, Alexander et al. identified single-nucleotide variants (SNVs) in growth factor genes associated with QOL in prostate cancer patients (5). Young et al. identified SNVs neural signalling genes associated with QOL and symptom burden in women undergoing cancer treatment (9). However, SNVs have not been investigated in the area QOL for patients receiving palliative radiotherapy for bone metastases. Our study aimed to identify candidate SNVs that are genetic molecular markers associated with changes in global QOL and pain in these patients.
Methods
Patient population
This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094). Patients provided informed consent before enrolling in the double-blind randomized NCIC Clinical Trials Group (NCIC CTG) Symptom Control 23 (SC. 23) study that assessed the effect of prophylactic dexamethasone on reducing pain flare in patients who were receiving palliative radiotherapy for bone metastases (10). Patients recorded their pain levels through completing the brief pain inventory (BPI), which ranked pain from a scale of 0 (no pain) to 10 (worst possible pain), and analgesic intake from day of radiation therapy (RT), then every day for 10 days post-RT, and at day 42 post-RT. Patients who reported a pain score of at least 2 at the treated site were eligible to participate in the study.
Survey analysis
To assess QOL, patients completed the European Organization of Cancer Research and Treatment (EORTC) QOL Questionnaire Core-15 Palliative (QLQ-C15-PAL) within 10 days of randomization (baseline), then at day 10 and 42 after RT. Patients with complete QOL-C15-PAL data were analysed. The QLQ-C15-PAL is a validated tool designed to assess QOL specifically in the palliative population. It comprises of 15 questions adapted from the QLQ-C30 core questionnaire. Responses to the global QLQ and pain items from the QOL-C15-PAL questionnaire were analysed. Both questions utilised a Likert scale, but while the QOL item was scored from 1 (very poor) to 7 (excellent), the pain item was scored from 1 (not at all) to 4 (very much). Therefore, higher scores represent better QOL but higher degree of pain (11,12). We then performed a linear transformation to obtain scores on a scale from 0-100. The differences between baseline and day 42 were used to calculate improvement or deterioration in QOL and pain, with positive differences representing improvement in QOL but worsening pain. Patients with absolute differences between baseline and day 42 that were greater than 10 were considered clinically significant and were analysed to determine genomic associations with change in global QOL and pain.
Genomic analysis
Patients provided saliva samples at day of RT, which was subsequently sequenced by the Illumina TruSightTM One Panel for 4,813 genes known to harbour disease-causing variants. BWA was used to map the raw sequencing data from Illumina’s MiSeq platform hg19 onto a reference genome (13). Base quality score recalibration, indel realignment, duplicate removal, and variant calling were performed based on GATK Best Practices to identify single nucleotide variants in the sequenced genes (14). Finally, ANNOVAR was used to annotate variants that had functional or clinical relevance, and to assist in variant filtering and analysis (15).
Variant filtering and selection
Variants from genes implicated in inflammation, radiation response, immune response, DNA damage, and QOL were selected for further analysis. On these variants, the Cochran-Armitage trend test was performed to look for significant associations with change in QOL and pain. Variants that had a significance level of P<0.005 underwent backwards elimination using the HPGENSELECT procedure on SAS in combination with the LASSO method of variable selection using the minimum Bayesian information criterion. The generated multivariable model with estimated effect sizes were used to calculate prognostic scores for QOL and for pain through multiplying the estimated effect size of each SNV with its corresponding SNV value. The calculated prognostic score of each patient was used to divide patients in to risk groups of low (<1/3 quartiles), medium (≥1/3 but <2/3 quartiles), and high (≥2/3 quartiles). Next, the Cochran-Armitage trend test was used for univariate analysis of the risk groups. For multivariable analysis, a logistic regression model was fitted to the risk group model and adjusted for gender and primary cancer site.
Variants selected in the multivariable model underwent literature search and pathway analysis to find existing associations and biological pathways that may be clinically or biologically relevant in the improvement or deterioration in QOL and pain.
Results
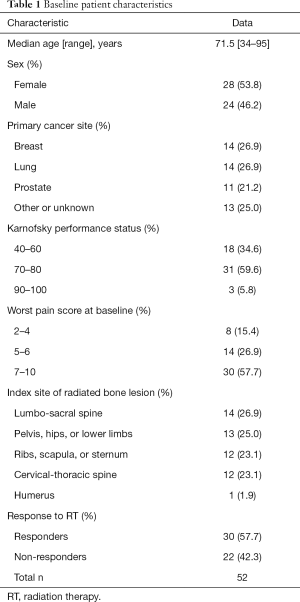
A total of 52 patients were analysed for global QOL. The baseline characteristics of these patients are presented in Table 1. Their median age was 71.5 years old (range 34–95 years), and 54% of patients were female. The most common primary cancer sites were breast (27%), lung (27%) and prostate (21%). Patients most frequently had a Karnofsky performance status of 70–80 (60%) and a high worst pain score of 7–10 at baseline (58%). Palliative radiation was most commonly prescribed to the lumbo-sacral spine (27%) followed by pelvis, hip, or lower limbs (25%), Cervical-thoracic spine (23%) and Ribs, clavicle or sternum (23%). Out of 52 patients, 30 (58%) responded to RT.
Full table
Variants associated with the C15-PAL global QOL score
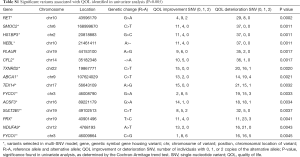
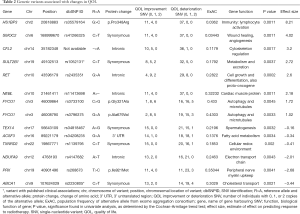
Univariate analysis found 15 variants significantly associated with change in QOL (Table S1). The most significant variant was the rs2435351 G>A intronic variant from the gene RET (P=0.0002). Fourteen of these variants were selected in the multivariable model (Table 2). The variant with the largest effect size was the rs35579164 G>C variant of the gene HS1BP3, corresponding to an amino acid change from proline to arginine at position 348 (effect size =8.21, P=0.0011). The variant with the largest negative effect size was the rs2230805 synonymous variant from the gene ABCA1 (effect size =−3.44, P=0.0021). One gene, FYCO1, contributed two variants to the multivariable model. These are the rs3796375 G>A variant which produces an amino acid change from alanine to valine at position 679 (effect size =1.02, P=0.0033), and the rs3733100 C>G variant which produces an amino acid change from glycine to alanine at position 321 (effect size =1.72, P=0.0045).
Full table
Full table

Full table
Univariate analysis of the risk groups derived from the multivariable model found that risk groups were highly predictive of a patient’s global QOL response to radiotherapy (P<0.0001, Table 3). In multivariable analysis with logistic regression and accounting for preselected baseline factors, the risk groups remained significantly predictive of pain response to radiotherapy [OR: 2.1, 95% confidence interval (CI): 1.2–3.9, P=0.015]. In univariate analysis, QOL response was not significantly associated with pain flare after radiotherapy (P=0.65), and remained insignificant after adjusting for baseline factors (OR: 1.2; 95% CI: 0.6–2.2, P=0.66).
Two variants were identified in the literature search. This included rs2230805 C>T from ABCA1, which produces a membrane transporter of lipids such as cholesterol. Several studies have investigated the role of rs2230805 and Alzheimer’s disease, and found that this variant was significantly associated with disease risk, possibly through influencing the lipid profile (16-19). A study by Pasdar et al. investigated the role of rs2230805 and ischemic stroke, but found no associations (20). The second variant was rs1052131 from the gene SULT2B1, which produces a protein involved in the metabolism of steroid compounds. The variant allele was found to be associated with a reduced risk of esophageal squamous cell carcinoma (21).
Variants associated with the C15-PAL pain scale score
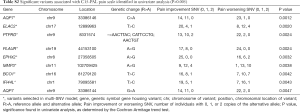
We identified 9 variants significantly associated with pain in univariate analysis (Table S2). The most significant variant was rs76608797 C>A of the aquaporin gene AQP7, which produces an amino acid change from valine to phenylalanine at position 152 (P=0.0012). This gene had a second variant significant in univariable analysis, a G>A variant at position 33386144 (P=0.0047). Eight of these variants were selected in the multivariable model (Table 4). The variant with the largest positive effect size was rs4760 A>G from the gene PLAUR, which produces a protein change from leucine to proline at position 193 (effect size =5.23, P=0.0024). The variant with the largest negative effect size was rs11545302 T>C, a synonymous variant of the gene ELAC2 (effect size =−0.84, P=0.002).
Full table
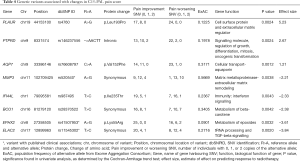
Full table
Univariate analysis of the multi-SNV risk group model found that the risk groups were highly predictive of a patient’s pain response to radiotherapy (P<0.0001, Table 5). However, the risk group did not predict pain response in logistic model adjusted for age and primary cancer site- (OR: 0.93; 95% CI: 0.5–1.6, P=0.79).

Full table
Three of the variants in the model have been published. Beuten et al. found that rs11545302 of ELAC2, a gene that produces a protein involved in tRNA processing, transforming growth factor-beta (TGF-β) signalling, and the DNA damage response pathway, was significantly associated with prostate cancer risk (22). This was especially significant in Caucasian men, in a manner independent from other significant SNVs they identified (22). The rs41507953 A>G variant of EPHX2 produces an amino acid change from lysine to arginine at position 55. Gervasini et al. studied three variants of EPHX2 in renal transplant recipients and found that there was a trend of the haplotype rs41507953 A/rs751141 A/rs1042032 G being associated with a higher risk of acute rejection episodes, but it did not reach statistical significance (P=0.08) (23). Rs520540 A>G is a synonymous exonic variant of the gene MMP3, which produces a matrix metalloproteinase that remodels the extracellular matrix. This SNV has been investigated for association with sporadic Alzheimer’s disease and ischemic stroke in multiple studies, but with inconclusive evidence (24-28). Matarin et al. studied the association between this SNV and ischemic stroke risk, and found a significant association in the discovery phase (P=0.02) but no significance in upon replication (P=0.68) (26). Shibata et al. found that rs520540 did not influence the risk of Alzheimer’s disease (28).
Discussion
Our study identified a predictive multi-SNV model associated with clinically significant changes in global QOL or pain in patients with bone metastases who underwent palliative radiation. Cytochrome proteins are involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. For example, CYP7B1 expression is regulated by androgen and estrogens, and its activity is associated with cellular proliferation (29). Furthermore, studies in rat hepatocytes showed that application of steroid dexamethasone increased CYP7B1 activity (30). In the present study, dexamethasone has been prescribed to a subset of the patient population as part of the primary objective of the SC23 trial that identified its efficacy in prophylaxis of radiation-induced pain flare of patients receiving palliative radiation to bone metastases (10). While variants of cytochromes have not been identified, our model identified significant variants from other genes involved in lipid regulation, metabolism and lipid transport. This includes a cholesterol transporter (ABCA1), and enzymes that metabolise fatty acids (ACSF3), beta-carotene (BCO1), and epoxides (EPHX2). Therefore, further investigation and validation of SNVs in these genes are required to clarify their potential roles in controlling dexamethasone response, QOL, and response to palliative radiotherapy.
One of the SNVs identified in our model was from the gene PRX, which produces periaxin, a protein that is directly involved influencing pain signalling through stabilizing the peripheral nervous system myelin. Animal studies have found that individuals deficient in periaxin have unstable myelin sheaths, demyelination, and behaviours associated with peripheral nerve damage such as mechanical allodynia and thermal hyperalgesia (31). Clinically, patients with mutations in PRX also display neuropathy (32). Therefore, the novel rs268673 T:C PRX SNV we identified may influence pain signalling, and through this, affect a patient’s QOL.
QOL encompasses multiple functional and symptomatic domains of life such as nausea and vomiting, fatigue, and pain (3). Therefore, the potential for genetic biomarkers obtained non-invasively through saliva samples to predict global QOL and pain enables targeted management in these individuals. Our study was limited by small sample size. Further investigation is required to validate the candidate SNVs identified in this study in order to establish a set of genetic biomarkers capable of predicting patient QOL in cancer patients with bone metastases after receiving palliative radiotherapy.
Acknowledgements
We thank the generous support of Joey and Mary Furfari Cancer Research Fund. We thank Dr. Ralph Meyer for his contributions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094). Written informed consent was obtained from the patient for publication of this manuscript.
References
- Cai B, Nickman NA, Gaffney DK. The role of palliative external beam radiation therapy in boney metastases pain management. J Pain Palliat Care Pharmacother 2013;27:28-34. [Crossref] [PubMed]
- Shibata H, Kato S, Sekine I, et al. Diagnosis and treatment of bone metastasis: comprehensive guideline of the Japanese Society of Medical Oncology, Japanese Orthopedic Association, Japanese Urological Association, and Japanese Society for Radiation Oncology. ESMO Open 2016;1:e000037. [Crossref] [PubMed]
- Caissie A, Culleton S, Nguyen J, et al. EORTC QLQ-C15-PAL quality of life scores in patients with advanced cancer referred for palliative radiotherapy. Support Care Cancer 2012;20:841-8. [Crossref] [PubMed]
- Katz N. The Impact of Pain Management on Quality of Life. J Pain Symptom Manage 2002;24:S38-47. [Crossref] [PubMed]
- Alexander KE, Chambers S, Spurdle AB, et al. Association between single-nucleotide polymorphisms in growth factor genes and quality of life in men with prostate cancer and the general population. Qual Life Res 2015;24:2183-93. [Crossref] [PubMed]
- Schoormans D, Radonic T, de Witte P, et al. Mental Quality of Life Is Related to a Cytokine Genetic Pathway. PLoS One 2012;7:e45126. [Crossref] [PubMed]
- Sprangers MA, Sloan JA, Veenhoven R, et al. The Establishment of the GENEQOL Consortium to Investigate the Genetic Disposition of Patient-Reported Quality-of-Life Outcomes. Twin Res Hum Genet 2009;12:301-11. [Crossref] [PubMed]
- Lupski JR. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ Mol Mutagen 2015;56:419-36. [Crossref] [PubMed]
- Young EE, Kelly DL, Shim I, et al. Variations in COMT and NTRK2 influence symptom burden in women undergoing breast cancer treatment. Biol Res Nurs 2017;19:318-28. [Crossref] [PubMed]
- Chow E, Meyer RM, Ding K, et al. Dexamethasone in the prophylaxis of radiation-induced pain flare after palliative radiotherapy for bone metastases: a double-blind, randomised placebo-controlled, phase 3 trial. Lancet Oncol 2015;16:1463-72. [Crossref] [PubMed]
- Groenvold M, Petersen MA, Aaronson NK, et al. The development of the EORTC QLQ-C15-PAL: A shortened questionnaire for cancer patients in palliative care. Eur J Cancer 2006;42:55-64. [Crossref] [PubMed]
- Chow E, Hird A, Velikova G, et al. The European Organisation for Research and Treatment of Cancer Quality of Life Questionnaire for patients with Bone Metastases: The EORTC QLQ-BM22. Eur J Cancer 2009;45:1146-52. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Wolf A, Bauer B, Hartz AM. ABC Transporters and the Alzheimer’s Disease Enigma. Front Psychiatry 2012;3:54. [Crossref] [PubMed]
- Reynolds CA, Hong MG, Eriksson UK, et al. Analysis of lipid pathway genes indicates association of sequence variation near SREBF1/TOM1L2/ATPAF2 with dementia risk. Hum Mol Genet 2010;19:2068-78. [Crossref] [PubMed]
- Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochim Biophys Acta 2010;1801:824-30. [Crossref] [PubMed]
- Reynolds CA, Hong MG, Eriksson UK, et al. A survey of ABCA1 sequence variation confirms association with dementia. Hum Mutat 2009;30:1348-54. [Crossref] [PubMed]
- Pasdar A, Yadegarfar G, Cumming A, et al. The effect of ABCA1 gene polymorphisms on ischaemic stroke risk and relationship with lipid profile. BMC Med Genet 2007;8:30. [Crossref] [PubMed]
- Hyland PL, Freedman ND, Hu N, et al. Genetic variants in sex hormone metabolic pathway genes and risk of esophageal squamous cell carcinoma. Carcinogenesis 2013;34:1062-8. [Crossref] [PubMed]
- Beuten J, Gelfond JA, Franke JL, et al. Single and multivariate associations of MSR1, ELAC2, and RNASEL with prostate cancer in an ethnic diverse cohort of men. Cancer Epidemiol Biomarkers Prev 2010;19:588-99. [Crossref] [PubMed]
- Gervasini G, Garcia-Cerrada M, Coto E, et al. A 3’-UTR Polymorphism in Soluble Epoxide Hydrolase Gene Is Associated with Acute Rejection in Renal Transplant Recipients. PLoS One 2015;10:e0133563. [Crossref] [PubMed]
- Kim SK, Kang SW, Kim DH, et al. Matrix Metalloproteinase-3 Gene Polymorphisms Are Associated with Ischemic Stroke. J Interferon Cytokine Res 2012;32:81-6. [Crossref] [PubMed]
- Chang JJ, Stanfill A, Pourmotabbed T. The role of matrix metalloproteinase polymorphisms in ischemic stroke. Int J Mol Sci 2016.17. [PubMed]
- Matarin M, Brown WM, Dena H, et al. Candidate gene polymorphisms for ischemic stroke. Stroke 2009;40:3436-42. [Crossref] [PubMed]
- Heikkinen AM, Raivisto T, Kettunen K, et al. Pilot Study on the Genetic Background of an Active Matrix Metalloproteinase-8 Test in Finnish Adolescents. J Periodontol 2017;88:464-72. [Crossref] [PubMed]
- Shibata N, Ohnuma T, Higashi S, et al. Genetic association between matrix metalloproteinase MMP-9 and MMP-3 polymorphisms and Japanese sporadic Alzheimer’s disease. Neurobiol Aging 2005;26:1011-4. [Crossref] [PubMed]
- Tang W. Hormonal Regulation of the Human CYP27A1 and CYP7B1 Genes. Uppsala: Acta Universitatis Upsaliensis, 2007.
- Pandak WM, Hylemon PB, Ren S, et al. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in primary cultures of rat hepatocytes. Hepatology 2002;35:1400-8. [Crossref] [PubMed]
- Gillespie CS, Sherman DL, Fleetwood-Walker SM, et al. Peripheral demyelination and neuropathic pain behavior in periaxin-deficient mice. Neuron 2000;26:523-31. [Crossref] [PubMed]
- Marchesi C, Milani M, Morbin M, et al. Four novel cases of periaxin-related neuropathy and review of the literature. Neurology 2010;75:1830-8. [Crossref] [PubMed]


