Genetic biomarkers associated with pain flare and dexamethasone response following palliative radiotherapy in patients with painful bone metastases
Introduction
Radiation therapy (RT) is commonly used in palliation of painful bone metastases. However, a transient increase in pain within 10 days since treatment known as a “pain flare” is experienced by around 40% of patients who receive palliative radiotherapy (1). As radiation is inherently cytotoxic, it may cause pain flare by evoking an endogenous inflammatory response. Analysis of levels of urinary inflammatory proteins showed an overall increase in levels of cytokines and chemokines following radiotherapy. Patients who experienced pain flare also had different urinary protein profiles compared to patients who did not experience pain flare. These patients had significantly lower levels of pro-inflammatory chemokines interleukin-8 (IL-8), IL-10, and macrophage derived chemokine (MDC) (2).
It has been postulated that the imbalance of pro-inflammatory and anti-inflammatory cytokines contributes to the development of pain flare. Dexamethasone is prophylactically prescribed for managing side effects from radiation of brain metastases. A phase 3 trial conducted across 23 Canadian cancer centers found that dexamethasone is also effective in reducing the incidence of pain flare from 35% to 26% after palliative radiation to bone metastases (3). It has been proposed that differences in dexamethasone response are due to variations in metabolism among individuals, which may also be observed at a genetic level.
Genetic biomarkers have been used to identify individual propensities in treatment response. For example, 58–78% of the individual differences in sensitivity to radiotherapy can be attributed to heritable genetic factors (4). Many of these identified genetic differences are single nucleotide variations (SNVs) of genes involved in DNA damage response, inflammation, and growth factor signalling pathways. For example, the SNV rs12757998 from the gene RNASEL that encodes for a pro-inflammatory ribonuclease has been found to be associated with outcome in prostate cancer patients who received RT (5). Our study aimed to identify genetic biomarkers associated with pain flare and dexamethasone response. This would enable individualised treatment plans for more effective management of pain in patients with bone metastases.
Methods
Patient population
This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094). Written informed consent was obtained for cancer patients at the Odette Cancer Centre receiving a single 8 Gy dose of palliative RT for painful bone metastases before enrollment into the multi-centre NCIC Clinical Trials Group (NCIC CTG) Symptom Control 23 (SC.23) study (3). Patients were randomized into one of two arms: prophylactic dexamethasone (two 4 mg tablets) taken at least 1 hour before RT and for 4 days after RT, or placebo pills for control.
Data collection
To assess pain flare and dexamethasone response, patients filled out the brief pain inventory (BPI) and recorded their analgesic intake on day of RT, every day for 10 days post-RT, and at day 42 post-RT. The BPI ranked pain on a scale of 0 to 10, with 0 being no pain, and 10 being pain as bad as you can imagine. Analgesic intake was converted into daily oral morphine equivalent. Pain flare was defined as at least a two-point increase in the worst pain score without reduction in analgesic intake, or at least a 25% increase in analgesic intake without worst pain score reduction. To be categorized as pain flare rather than pain progression, pain score and analgesic intake must have returned to baseline during the 10 days after RT. Dexamethasone response was determined by whether patients had pain flare despite having taken prophylactic dexamethasone.
Genomic analysis
Saliva samples were taken at day of RT, and underwent DNA sequencing using the Illumina TruSightTM One Panel. This identified SNVs in 4,813 genes harbouring disease-causing variants. Burrows-Wheeler Aligner’s Smith-Waterman alignment (BWA) was used to map the raw sequencing data from Illumina’s MiSeq platform hg19 to a reference genome (6). As outlined by Genome Analysis Tool Kit (GATK) Best Practice, we performed base quality score recalibration, indel realignment, duplicate removal, and variant calling (7).
Variant selection
Genes that were associated with inflammation, radiation response, immune response, or DNA damage were selected for variant analysis. The Cochran-Armitage trend test was used to assess associations between variants and pain flare or pain flare treated with dexamethasone.
For the multivariable model predictive of pain flare or pain flare treated with dexamethasone, statistically significant variants with P<0.005 underwent backwards elimination. The hpgenselect procedure on SAS was applied using the LASSO method of variable selection with the minimum Bayesian Information Criterion. The prognostic scores for pain flare were produced from the sum of the estimate of effect in the hpgenselect model of each of SNVs in the multivariable model, multiplied by the corresponding SNV value. The prognostic scores were calculated for each patient and used to divide patients into risk groups of low (<1/3 quartiles), medium (≥1/3 quartiles but <2/3 quartiles) and high (≥2/3 quartiles). Univariate analysis of the risk groups was conducted using the Cochran-Armitage trend test. For multivariable analysis of pain flare, a logistic regression model was generated, using pain flare as outcome (low risk of pain flare as the referent group), and baseline factors (gender and primary cancer type)-adjusted risk group model of pain flare as the independent factor.
Univariable analysis of pain flare after treated with dexamethasone was conducted in patients who received dexamethasone. Variants were then selected from the significant variants for the multivariable model and prognostic score, which was used to calculate risk scores of patients in both arms of dexamethasone and placebo. Patients from each arm were then divided into three risk groups. Data from both arms were combined to produce a logistic regression model with pain flare as the outcome variable, fit with risk groups, treatment arm, and interaction terms, adjusted for gender and primary cancer site.
The identified variants in the prognostic model underwent pathway analysis and literature search to identify potential biological mechanisms associated with pain flare or dexamethasone response.
Results
A total of 81 patients were included in biomarkers analysis, which consisted of 50 patients who received prophylactic dexamethasone and 31 patients who did not receive prophylactic dexamethasone. The median patient age was 72 (range, 33–95) years old, and 56% of patients were male (Table 1). The most common primary cancer sites were prostate (32%), followed by breast (24%) and lung (24%). The majority of patients had a Karnofsky performance status between 70–80 (62%), and a worst pain score at baseline of 7–10 (56%). Radiation to the pelvis, hips, or lower limbs was most common (35%). Twenty-two (27%) patients experienced pain flare. In 50 patients who took prophylactic dexamethasone, 11 (22%) had pain flare.
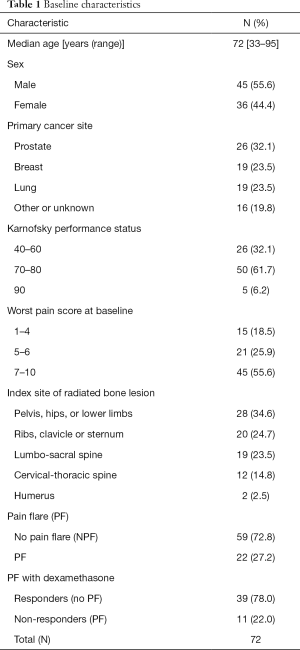
Full table
Variants associated with pain flare
In the univariate analysis, we identified 50 variants associated pain flare (Table S1). The most significant variants associated with pain flare were the A>C variant of DNM2 at position 10940838 (P=0.0002), and the G>A variant of UGT2A1-UGT2A2 at position 70505162 (P=0.0002).
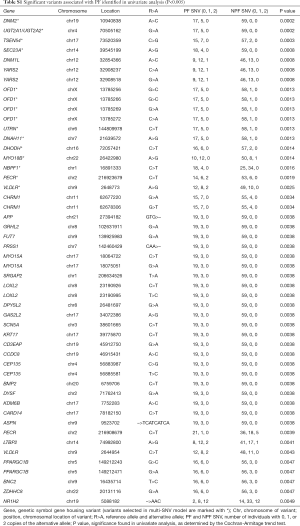
Full table
Fifteen variants were selected in the multivariable model (Table 2). Individuals with high prognostic scores belonged to higher risk groups, which were associated with the development of pain flare. In univariate analysis, the risk groups were significantly predictive of pain flare (P<0.0001), with patients in the high-risk group patients having a 78% chance of developing pain flare (Table 3).
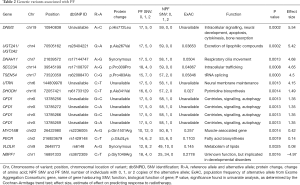
Full table
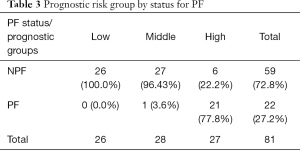
Full table
In the multivariable model, the variant with the largest positive effect size was also one of the most significant variants identified in the univariate analysis, namely the A>C variant at position 10940838 of DNM2 (effect size =5.54). This variant produced an amino acid change from histidine to leucine at position 772. One variant had a negative effect size. This was rs3872309 of NBPF1, which had a C>T variant at position 16891333 that produced an amino acid change from glycine to arginine at position 1049 (effect size =−4.97, P=0.00016).
The X-chromosome gene OFD1 had four variants in the multivariable model. These were the G>C variant at position 13785256 and 13785266, the C>A variant at position 13785272, and the G>A variant at position 13785269. All four variants had an effect size of 1.35, and P=0.0013 in univariate statistical analysis.
Variants associated with pain flare despite dexamethasone
In univariate analysis, we identified 25 variants with P<0.005 that were associated with pain flare with dexamethasone (Table S2). Among these variants, the most significant were the G>A variant of CREBBP at position 3795363 (P=0.0007), and the G>A variant of HAS1 at position 52220351 (P=0.0007).
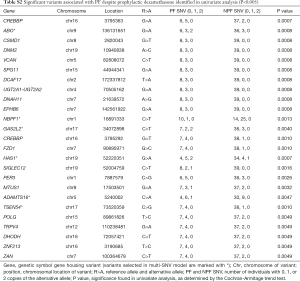
Full table
The multivariate model included six variants, with their effect size and statistical significance from the univariate model shown on Table 4. High prognostic scores are predictive of developing pain flare for patients on dexamethasone (P<0.0001, Table 5). Patients who were on dexamethasone were also significantly less likely to develop pain flare (P=0.023). After adjustment for the preselected baseline factors, the high-risk group remained significantly predictive of pain flare after radiotherapy in patients treated with dexamethasone [OR =24.6; 95% confidence interval (CI) 1.8–342.7, P=0.02].

Full table
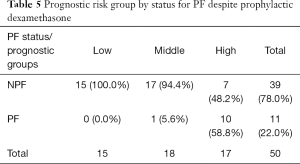
Full table
The SNV with the largest effect size was rs62088470 of TSEN54, a C>G variant at position 73520359 that produced an amino acid change from proline to alanine at position 483 (effect size =0.85, P=0.0010). One SNV had a negative effect size. This was the rs3872309 variant of NBPF1, corresponding to a C>T variant at position 16891333 that produces an amino acid change from glycine to arginine at position 1049 (effect size =−1.03, P=0.0013).
Variants with pre-existing clinical investigations from literature review
Our literature review identified none of the genetic variants in the model predicting pain flare. However, two SNVs in the model predicting dexamethasone response have been published. This includes rs1053878 of the gene ABO that corresponds a G>A variant at position 136131651 which produces an amino acid change from proline to leucine at position 156. Through a retrospective cohort study, Cozzi et al. identified that ovarian cancer patients who are minor allele carriers of rs1053878 had better overall survival (OS) (8). This corresponds to improved survival in patients with blood type A, when compared to those with blood types B or O.
This also includes rs11084111, a synonymous variant of a G>A change at position 52220351 of HASI, a gene that produces the extracellular matrix compound hyaluronic acid. Bulatova et al. studied whether polymorphisms of rs11084111 affected disease progression in patients with chronic hepatitis C (9). However, the authors found no significant difference between rs11084111 alleles among healthy patients compared to patients with chronic hepatitis C.
Discussion
We identified SNVs that belong to several significant pathways, including those of biosynthesis, metabolism, excretion, and intracellular signalling. These pathways may be important in the mechanisms underlying variation among patients in experiencing pain flare and in response to dexamethasone.
Pain flare has been proposed to be due to an inflammatory response from certain individuals in response to cell death induced by radiotherapy (10). One of the genes implicated in pain flare in our model is DNM2, which produces the protein dynamin. Dynamin is an intracellular signalling protein that is member of the GTPase family, and is involved in a variety of cellular processes including apoptosis (11). Therefore, variation in this gene may lead to differences in the cellular response to radiation and subsequent inflammatory response.
Metabolic genes and genes involved in excretion had been identified as significantly associated with pain flare. For instance, rs28404221 was identified in the genes UGT2A1-UGT2A2 of the UDP glucuronosyltransferase 2 family, which produce proteins that catalyze the conjugation of lipophilic substrates with glucuronic acid to increase water solubility and enhance excretion (12). UGT2A1 is known as a detoxification enzyme, and some of its polymorphisms have been identified as playing potential roles in tobacco-associated carcinogenesis (12).
Another gene involved in metabolism is VLDLR, which produces a transmembrane receptor for very-low-density lipoproteins (VLDLs). Our study identified that one of its synonymous variants, rs6148, is in a model that may be predictive of pain flare. VLDLR is important in cholesterol homeostasis, and therefore may be contribute to the physiological response to dexamethasone, which, like cholesterol, is a member of the steroid family (13). Moreover, it has been suggested that VLDLR is involved in inducing adipose tissue inflammation (14). Therefore, variation in VLDLR may also contribute to the development of pain flare.
Being a synthetic steroid, dexamethasone exerts its physiological effects through binding and activating glucocorticoid receptors located in the nucleus, leading to transcription of genes that produce an anti-inflammatory response (15,16). Our panel included four glucocorticoid receptor genes: NR3C1, NR3C2, NR4A2, and NR5A1, but none yielded significant variants. Since the downstream anti-inflammatory effects of dexamethasone are multifaceted, and include suppressing prostaglandin synthesis, and transcription of pro-inflammatory interleukins such as IL-1β, IL-6, and Il-8, it is possible that the variation among how individuals respond to dexamethasone may be due to differences in how these pathways are influenced by it. Our study, limited by small sample size, at the most can serve as hypotheses generating and would require validation in future larger research studies.
Finding a panel of variants that can predict pain flare and dexamethasone response is of great clinical utility. With the rapid advancement of genetic technology in terms of efficiency and speed, genetic methods will likely become more accessible and affordable in identifying patient responses to treatment options and enable more effective care. As the genomic data of a patient are stable across time, gene assessment may also be conducted well in advance of the delivery of palliative care. This can reduce patient burden by eliminating treatments that are not likely to be beneficial. Therefore, further study is required to independently validate our proposed predictive genetic models. This would enable the identification of individuals who are at risk of pain flare, and therefore allow intervention before the onset of increased pain after palliative radiotherapy. This can also aid in the identification of an appropriate prophylactic medication, based on whether an individual will respond to dexamethasone. If not, analysis and further study of the non-responders may enable identification of alternative measures to prevent pain and improve well-being.
Acknowledgements
We thank the generous support of the Joey and Mary Furfari Cancer Research Fund. We thank Dr. Ralph Meyer for his contributions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Ontario Cancer Research Ethics Board (OCREB) (No. 10-094). Written informed consent was obtained for cancer patients at the Odette Cancer Centre.
References
- McDonald R, Chow E, Rowbottom L, et al. Incidence of pain flare in radiation treatment of bone metastases: A literature review. J Bone Oncol 2014;3:84-9. [Crossref] [PubMed]
- Bushehri A, Chow E, Zhang L, et al. Urinary cytokines/chemokines pattern in patients with painful bone metastases undergoing external beam radiotherapy experiencing pain flare. Ann Palliat Med 2016;5:107-15. [Crossref] [PubMed]
- Chow E, Meyer RM, Ding K, et al. Dexamethasone in the prophylaxis of radiation-induced pain flare after palliative radiotherapy for bone metastases: a double-blind, randomised placebo-controlled, phase 3 trial. Lancet Oncol 2015;16:1463-72. [Crossref] [PubMed]
- Guo Z, Shu Y, Zhou H, et al. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis 2015;36:307-17. [Crossref] [PubMed]
- Schoenfeld JD, Margalit DN, Kasperzyk JL, et al. A single nucleotide polymorphism in inflammatory gene RNASEL predicts outcome after radiation therapy for localized prostate cancer. Clin Cancer Res 2013;19:1612-9. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Cozzi GD, Levinson RT, Toole H, et al. Blood type, ABO genetic variants, and ovarian cancer survival. PLoS One 2017;12:e0175119. [Crossref] [PubMed]
- Bulatova IA, Schekotova AP, Krivtsov AV, et al. The noninvasive evaluation of degree of expression of fibrosis of liver and significance of polymorphism of gene of hyaluronic acid under chronic hepatitis C. Klin Lab Diagn 2015;60:18-21. [PubMed]
- Chow E, Ling A, Davis L, et al. Pain flare following external beam radiotherapy and meaningful change in pain scores in the treatment of bone metastases. Radiother Oncol 2005;75:64-9. [Crossref] [PubMed]
- Li J, Xu L, Ye J, et al. Aberrant dynamin 2-dependent Na(+) /H(+) exchanger-1 trafficking contributes to cardiomyocyte apoptosis. J Cell Mol Med 2013;17:1119-27. [PubMed]
- Bushey RT, Chen G, Blevins-Primeau AS, et al. Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet Genomics 2011;21:55-65. [Crossref] [PubMed]
- Go GW, Mani A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med. 2012;85:19-28. [PubMed]
- Nguyen A, Tao H, Metrione M, et al. Very low density lipoprotein receptor (VLDLR) expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J Biol Chem 2014;289:1688-703. [Crossref] [PubMed]
- Kil SH, Kalinec F. Expression and dexamethasone-induced nuclear translocation of glucocorticoid and mineralocorticoid receptors in guinea pig cochlear cells. Hear Res 2013;299:63-78. [Crossref] [PubMed]
- Newton R. Molecular mechanisms of glucocorticoid action: what is important? Thorax 2000;55:603-13. [Crossref] [PubMed]


